Yeast Sphingolipid Metabolism
The Author: Robert C Dickson, Professor of Molecular and Celliular Biochemistry,University of Kentucky College of Medicine, Lexington, KY, USA 40506-9983
Introduction
Sphingolipids along with sterols and glycerophospholipids are the lipid building blocks or structural lipids for constructing membranes in eukaryotic cells. In addition, some members of each of these lipid classes serve as signaling molecules to regulate biological processes. Some lipids acting as signals reside in membranes while others have a non-membrane location [1, 2].
A great many species of yeasts inhabit planet earth, but sphingolipids have been characterized in only a few of these. In contrast, sphingolipids have been extensively characterized in the yeast Saccharomyces cerevisiae, known as a budding yeast because of the way in which it reproduces. S. cerevisiae is also known as baker’s and brewing yeast because of its ability to metabolize sugars into carbon dioxide or ethanol, respectively. S. cerevisiae is commonly referred to as “yeast” or “yeasts” in the published literature.
There are several reasons why sphingolipids are best characterized in S. cerevisiae starting with fact that they can be grown easily in small or large quantities, stored for long periods of time and Mendelian or molecular techniques can be used to analyze and manipulate their genes. S. cerevisiae was the first eukaryotic organism to have its entire genomic sequence determined and for which a nearly complete set of single-gene deletion mutants was available, both of which contributed to our current knowledge of sphingolipids in this organism [3, 4]. Additionally, the Saccharomyces Genome Database, SGD (www.yeastgenome.org) was established in the early 1990s and has been an invaluable resource for fostering biological discovery.
Biosynthesis of Yeast Sphingolipids
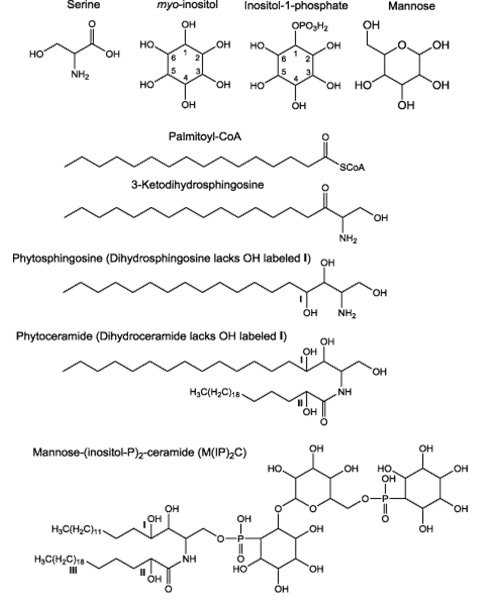
S. cerevisiae sphingolipids comprise about 7% of the lipid mass of the plasma membrane, where most sphingolipids are located. Another way of appreciating the abundance of sphingolipids in yeast cells is that they represent roughly thirty percent of all phospholipids which includes both sphingolipids and glycerophospholipids [5]. The chemical building blocks necessary for the synthesis of yeast sphingolipids are shown in Fig. 1.
Fig. 1. Chemical building blocks for yeast sphingolipids. The three sites of hydroxylation in yeast sphingolipids are indicated I, II or III. Hydroxylation at site I is catalyzed by the Sur2 enzyme using either dihydrosphingosine (DHS) or ceramide (phytosphingosine lacking hydroxylation at site II) as substrates as outlined on the metabolic pathway in Fig. 2. Sphingolipids phosphorylated at site II are referred to as α-hydroxylated [6]. Hydroxylation catalyzed by an unknown enzyme can occur also at site III on the long-chain fatty acid moiety of ceramides containing either dihydrosphingosine (DH-ceramides) or phytosphingosine (phyto-ceramides).
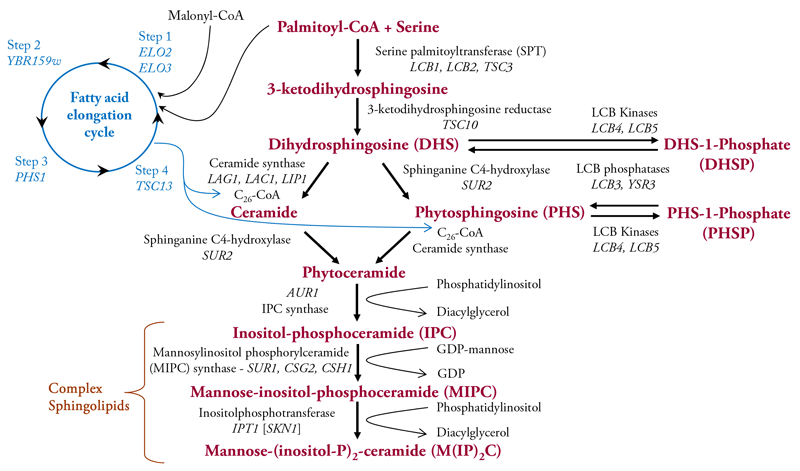
Enzymes that catalyze sphingolipid synthesis are all embedded in membranes probably because most of their substrates are hydrophobic, except serine and the polar head group’s phosphoinositol and mannose on the complex sphingolipids [6-11]. The first reaction in the de novo biosynthesis pathway (Fig. 2) condenses the amino acid serine with a fatty acid, typically C16 palmitic acid, attached to Coenzyme A (Palmitoyl-CoA) to produce 3-ketodihydrosphingosine. This reaction is followed by reduction to give dihydrosphingosine (DHS, also called sphinganine). DHS can be used as a substrate for three reactions: (i) N-acylation with a fatty acid to give ceramide; (ii) hydroxylation on carbon atom 4, by the Sur2 protein, to give phytosphingosine (PHS); (iii) and phosphorylation on the C1 hydroxyl to give DHS-1-phosphate (DHSP), a reaction catalyzed primarily be the LCB4 kinase and to a lesser extent by the LCB5 kinase. Hydroxylation of the C4 atom of DHS in ceramide to yield phytoceramide is catalyzed also by the Sur2 protein. This atom of is one of three carbons in yeast sphingolipids that can be hydroxylated. The other hydroxyls can be on the N-linked fatty acid at sites labelled II, catalyzed by the Sac7 protein, and III, enzyme unknown, as shown in Fig. 1. Lastly, PHS can be N-acylated with a fatty acid to yield phytoceramide or it can be phosphorylated to yield PHS-1-Phosphate (PHSP). DHSP and PHSP can be dephosphorylated by the LCB3 or YSR3 LCB phosphatases. DHS and PHS are typically referred to as long-chain bases because of their chemical structure.
Fig. 2. Diagram of sphingolipid metabolism in Saccharomyces cerevisiae primarily showing de novo synthesis reactions. Metabolic intermediates and the complex sphingolipids are indicated in bold maroon lettering while enzyme names genes are in black lettering with gene names indicated by italics. More information about genes and other features of yeast sphingolipid metabolism are available as SGD (www.yeastgenome.org ). The reaction cycle that produces the very long-chain C26 fatty-acid, characteristic of yeast sphingolipids, is indicated in blue.
All of the reactions comprising ceramide and phytoceramide biosynthesis occur in the endoplasmic reticulum as do the four reactions comprising the fatty acid elongation cycle [8, 9, 12, 13]. This cycle has four steps with the first involving a condensation of two carbon atoms from malonyl-CoA to an acyl-CoA, typically palmitoyl-CoA, to produce a 3-keto-intermediate. This reaction is catalyzed by Elo2 and Elo3. In the second step the keto group is reduced to a hydroxyl by the Ybr159w protein, followed by Phs1-catalyzed dehydration at step 3. The resulting enoyl is reduced by the Tsc13 enzyme in the fourth step to yield a C18-fatty acid which continues to be elongated so that eight more rounds of the elongation cycle yield a C26-fatty acid, referred to as a very-long chain fatty acid (VLCFA). Most phytoceramides contain a C26 fatty acid, but phytoceramides with C24 or C22 fatty acids are present at lower concentrations.
Ceramide and phytoceramide are transported from the endoplasmic reticulum to the Golgi apparatus by vesicle trafficking where the C1 hydroxyl is covalently linked to inositol phosphate to produce inositol-phosphoceramide (IPC, Fig. 2). The next reaction involves transfer of a mannose residue from GDP-mannose onto IPC to yield mannose-inositol-phosphoceramide (MIPC). In the terminal reaction a second inositol phosphate is added to MIPC to produce mannose-(inositol-P)2-ceramide M(IP)2C. The genes and enzymes for the steps in sphingolipid metabolism are indicated in Fig. 2 and more information can be found in the references listed below and at the SGD website.
The majority of sphingolipids made in the Golgi apparatus are transported to the plasma membrane by vesicle transport, but small amounts are transported to other membrane compartments including mitochondria [14-16]. The ratio of IPC, MIPC and M(IP)2C varies from strain to strain of S. cerevisiae. Other factors that can cause variation in the ratio of these complex sphingolipids are culture ingredients and other environmental conditions cells encounter along with growth phase (rapid dividing verses non-dividing cells). Typically M(IP)2C is the most abundant of the inositol-containing sphingolipids. Many types of sphingolipids in mammals are localized to one or the other leaflet of the plasma membrane but, contrary to what is often stated in the literature, this has not been determined for yeast sphingolipids.
To summarize the complexity of sphingolipids that can be made in yeast cells: Long-chain bases or their phosphorylated forms, ceramides and complex sphingolipids can contain either DHS or PHS. Furthermore, the ceramides and complex sphingolipids can contain or lack the hydroxyl at site labeled II (Fig. 1) or the alpha-carbon the DHS/PHS. And finally, a small fraction of ceramides and complex sphingolipids are hydroxylated at the site labeled III (Fig. 1).
Sphingolipids in other Yeast Species
S. cerevisiae only makes sphingolipids containing the characteristic inositolphosphoceramides in which ceramides are linked to a polar head group containing an inositol phosphate group. In contrast, many other eukaryotes including mammals and some yeasts make sphingolipids with ceramide linked to a sugar polar head group, glucose being the most common, to form the class of sphingolipids called glycosylceramides [17]. Many fungi make both classes of sphingolipids [18-20] as do plants[21]. Forests, especially oaks and other broadleaf trees and grapes are natural habitat for S. cerevisiae [22], so there is likely to be an evolutionary explanation for S. cerevisiae and other fungi have sphingolipids that are more plant-like than animal-like, since phosphoinositol-containing sphingolipids are not known to be present in higher animals including mammals.
Degradation and Turnover of Sphingolipids
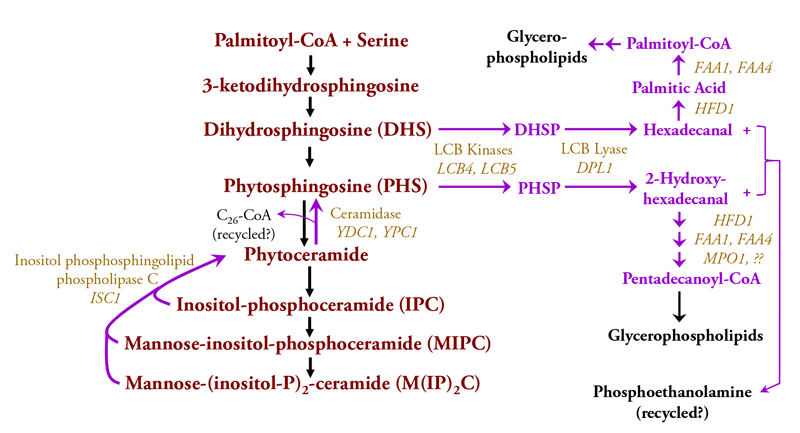
Most lipids in eukaryotic cell membranes are replaced by degradation and turnover mechanisms as a way for an organism to constantly react to changes in nutrients and other environmental factors. The known routes for degradation and turnover of S. cerevisiae sphingolipids are diagrammed in Fig. 3 [6-11].
Because complex sphingolipids (IPC, MIPC and M(IP)2C represent the bulk of sphingolipids in yeast cells, their degradation and turnover has been easier to characterize. However, our knowledge is limited and basic information such as the basal rate of sphingolipid turnover in growing or stationary phase (non-growing) yeast cells is not known. The initial step in the degradation of membrane-bound complex sphingolipids is catalyzed by the Isc1 enzyme (Inositol-phosphsphingolipid phospholipase C), which cleaves the bond between C1 of ceramide and the phosphoryl of the polar head group to yield phytoceramide and a free polar head group, either IP, MIP or M(IP)2. This type of cleavage is orthologous to the reaction catalyzed by mammalian neutral sphingomyelinase-2 that degrades sphingomyelin. The free polar head groups are probably recycled by a route that remains to be characterized. Phytoceramide is cleaved by a ceramides to yield a C26 fatty acid and PHS, which is phosphorylated on C1 to give PHS-1-Phosphate (PHSP) and then cleaved by the LCB lyase to produce the sixteen-carbon 2-hydroxyhexadecanal and phosphoethanolamine.
Only recently has the fate of the 2-hydroxyhexadecanal been characterized as outlined in Fig. 3[23]. A pathway with several steps converts 2-hydroxyhexadecanal, having an even number of carbon atoms, to the odd-numbered pentadecanoyl, which is converted to the CoA derivative and presumably recycled into glycerophospholipid metabolism as shown in Fig. 3.
Fig. 3. The pathways for degrading and turning over yeast sphingolipids are indicated by purple arrows. Enzymes that catalyzes degradation reactions are shown in light brown lettering and other coloring is the same as indicated in the legend to Fig. 2.
DHS can also be produced by turnover and degradation of complex sphingolipids containing ceramide (contains DHS) in place of phytoceramide (contains PHS). Such ceramide-containing sphingolipids are usually found at very low levels in wild-type cells but they are abundant in cells with a mutation in the SUR2 gene [24, 25] and they serve as substrates for the degradative enzymes shown in Fig. 3. DHS is phosphorylated by the same kinases as PHS to yield DHSP which is cleaved by the LCB lyase into hexadecanal and phosphoethanolamine. Hexadecanal can be recycled into glycerophospholipid metabolism [23].
Unanswered Questions about Yeast Sphingolipids
Yeast sphingolipids have been intensively studied over the past two decades and much has been learned because of advances in analytical techniques including fluorescent and other types of microscopy[26], large-scale analysis of mRNA levels [27], protein levels [28, 29] and post-translational modifications[30], and large-scale analysis of sphingolipids [31-33] and proteins that binding sphingolipids [34]. Systems biology, bioinformatics methods [33, 35-37] and network modeling [38, 39] have proven useful to integrate many types of data to better understand the cellular roles of yeast sphingolipids.
These studies have provided a wealth of information about yeast sphingolipids and revealed many cellular processes including or involving heat and salt stress, drug sensitivity, the endoplasmic reticulum, cell cycle control, the actin cytoskeleton, oxidative stress, aging and cell death pathways, protein turnover, nutrient sensing, cell wall metabolism, mitochondria and interactions with other lipids and lipid metabolism [6]. However, even these impressive efforts have yet to reveal how any individual sphingolipid modulates a signal transduction pathway or cellular processes in yeast.
There are several reasons why the functions of specific yeast sphingolipids is unknown with one reason being the complexity of sphingolipid metabolism, even in a single cell organism like yeast. Another reason is that sphingolipids can be rapidly converted into one or more intermediates in sphingolipid metabolism, a feature referred to as a “ripple effect” [36, 40]. A common way to understand how molecules work in biology is to genetically or pharmacologically block their function. This strategy works well for proteins because they typically interact in very specific ways with other molecules. However, this approach does not work well for sphingolipids because blocking an enzyme in sphingolipid metabolism causes ripple effects as intermediates on one side of the block tend to increase whereas ones on the other side tend to decrease. Thus, identifying which lipid intermediate is modulating a signal transduction pathway or a cellular process has proven to be nearly impossible.
Finally, it may be that single-cell organisms or less complex multicellular organisms, never required tight binding interactions between sphingolipids and proteins to produce biological effects. Such tight binding is a hallmark of several mammalian sphingolipids, the premier example being sphingosine-1-phosphate, which binds very tightly to a class of membrane-bound proteins called S1P receptors that regulate a wide array of biological processes [41]. Rather, yeast sphingolipids may primarily exert biological effects by using many weak interactions with proteins and or other membrane lipids.
References
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139-50. PubMed PMID: 18216770.
- Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep. 2004;5(8):777-82. PubMed PMID: 15289826.
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, et al. Life with 6000 genes. Science. 1996;274(5287):546-63.
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901-6. PubMed PMID: 10436161.
- Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J Bacteriol. 1991;173(10):3101-8.
- Montefusco DJ, Matmati N, Hannun YA. The yeast sphingolipid signaling landscape. Chemistry and physics of lipids. 2014;177:26-40. doi: 10.1016/j.chemphyslip.2013.10.006. PubMed PMID: 24220500; PubMed Central PMCID: PMC4211598.
- Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res. 2006;45(6):447-65. PubMed PMID: 16730802.
- Funato K, Vallee B, Riezman H. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry. 2002;41(51):15105-14. PubMed PMID: 12484746.
- Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol. 2010;688:217-31. Epub 2010/10/06. PubMed PMID: 20919657.
- Klug L, Daum G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014;14(3):369-88. doi: 10.1111/1567-1364.12141. PubMed PMID: 24520995.
- Breslow DK. Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb Perspect Biol. 2013;5(4):a013326. doi: 10.1101/cshperspect.a013326. PubMed PMID: 23545423; PubMed Central PMCID: PMC3683901.
- Breslow DK, Weissman JS. Membranes in balance: mechanisms of sphingolipid homeostasis. Mol Cell. 2010;40(2):267-79. Epub 2010/10/23. doi: S1097-2765(10)00781-1 [pii] 10.1016/j.molcel.2010.10.005. PubMed PMID: 20965421; PubMed Central PMCID: PMC2987644.
- Denic V, Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130(4):663-77. PubMed PMID: 17719544.
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192(1):7-16. doi: 10.1083/jcb.201006159. PubMed PMID: 21220505; PubMed Central PMCID: PMC3019561.
- Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146(4):741-54.
- Spincemaille P, Matmati N, Hannun YA, Cammue BP, Thevissen K. Sphingolipids and mitochondrial function in budding yeast. Biochim Biophys Acta. 2014;1840(10):3131-7. doi: 10.1016/j.bbagen.2014.06.015. PubMed PMID: 24973565; PubMed Central PMCID: PMC4145010.
- Merrill AH, Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev. 2011;111(10):6387-422. Epub 2011/09/29. doi: 10.1021/cr2002917. PubMed PMID: 21942574; PubMed Central PMCID: PMC3191729.
- Dickson RC, Lester RL. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426(2):347-57.
- Hannich JT, Umebayashi K, Riezman H. Distribution and functions of sterols and sphingolipids. Cold Spring Harb Perspect Biol. 2011;3(5). doi: 10.1101/cshperspect.a004762. PubMed PMID: 21454248; PubMed Central PMCID: PMC3101845.
- Rhome R, Del Poeta M. Lipid signaling in pathogenic fungi. Annu Rev Microbiol. 2009;63:119-31. doi: 10.1146/annurev.micro.091208.073431. PubMed PMID: 19450140.
- Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB. Plant sphingolipids: function follows form. Current opinion in plant biology. 2013;16(3):350-7. doi: 10.1016/j.pbi.2013.02.009. PubMed PMID: 23499054.
- Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23(9):494-501. PubMed PMID: 18656281.
- Kondo N, Ohno Y, Yamagata M, Obara T, Seki N, Kitamura T, et al. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nature communications. 2014;5:5338. doi: 10.1038/ncomms6338. PubMed PMID: 25345524.
- Shanklin J, Achim C, Schmidt H, Fox BG, Munck E. Mossbauer studies of alkane omega-hydroxylase: Evidence for a diiron cluster in an integral-membrane enzyme. Proc Natl Acad Sci U S A. 1997;94:2981-6.
- Grilley MM, Stock SD, Dickson RC, Lester RL, Takemoto JY. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273(18):11062-8. vNatter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, et al. The Spatial Organization of Lipid Synthesis in the Yeast Saccharomyces cerevisiae Derived from Large Scale Green Fluorescent Protein Tagging and High Resolution Microscopy. Mol Cell Proteomics. 2005;4(5):662-72. PubMed PMID: 15716577.
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278(5338):680-6. Epub 1997/10/24. PubMed PMID: 9381177.
- Dephoure N, Gygi SP. Hyperplexing: a method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Sci Signal. 2012;5(217):rs2. Epub 2012/03/30. doi: 5/217/rs2 [pii] 10.1126/scisignal.2002548. PubMed PMID: 22457332.
- Webb KJ, Xu T, Park SK, Yates JR, 3rd. Modified MuDPIT Separation Identified 4488 Proteins in a System-wide Analysis of Quiescence in Yeast. J Proteome Res. 2013;12(5):2177-84. Epub 2013/04/02. doi: 10.1021/pr400027m. PubMed PMID: 23540446.
- Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150(2):413-25. Epub 2012/07/24. doi: S0092-8674(12)00706-4 [pii] 10.1016/j.cell.2012.05.036. PubMed PMID: 22817900; PubMed Central PMCID: PMC3404735.
- Tarasov K, Stefanko A, Casanovas A, Surma MA, Berzina Z, Hannibal-Bach HK, et al. High-content screening of yeast mutant libraries by shotgun lipidomics. Mol Biosyst. 2014. doi: 10.1039/c3mb70599d. PubMed PMID: 24681539.
- Guan XL, Riezman I, Wenk MR, Riezman H. Yeast lipid analysis and quantification by mass spectrometry. Meth Enzymol. 2010;470:369-91. Epub 2010/10/16. doi: S0076-6879(10)70015-X [pii]10.1016/S0076-6879(10)70015-X. PubMed PMID: 20946818.
- Montefusco DJ, Chen L, Matmati N, Lu S, Newcomb B, Cooper GF, et al. Distinct signaling roles of ceramide species in yeast revealed through systematic perturbation and systems biology analyses. Sci Signal. 2013;6(299):rs14. doi: 10.1126/scisignal.2004515. PubMed PMID: 24170935.
- allego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol. 2010;6:430. Epub 2010/12/02. doi: msb201087 [pii]10.1038/msb.2010.87. PubMed PMID: 21119626; PubMed Central PMCID: PMC3010107.
- Montefusco DJ, Newcomb B, Gandy JL, Brice SE, Matmati N, Cowart LA, et al. Sphingoid bases and the serine catabolic enzyme CHA1 define a novel feedforward/feedback mechanism in the response to serine availability. J Biol Chem. 2012;287(12):9280-9. Epub 2012/01/27. doi: M111.313445 [pii]0.1074/jbc.M111.313445. PubMed PMID: 22277656; PubMed Central PMCID: PMC3308826.
- Cowart LA, Shotwell M, Worley ML, Richards AJ, Montefusco DJ, Hannun YA, et al. Revealing a signaling role of phytosphingosine-1-phosphate in yeast. Mol Syst Biol. 2010;6:349. Epub 2010/02/18. doi: msb20103 [pii]10.1038/msb.2010.3. PubMed PMID: 20160710; PubMed Central PMCID: PMC2835565.
- da Silveira Dos Santos AX, Riezman I, Aguilera-Romero MA, David F, Piccolis M, Loewith R, et al. Systematic lipidomic analysis of yeast protein kinase and phosphatase mutants reveals novel insights into regulation of lipid homeostasis. Mol Biol Cell. 2014;25(20):3234-46. doi: 10.1091/mbc.E14-03-0851. PubMed PMID: 25143408; PubMed Central PMCID: PMC4196872.
- Borklu Yucel E, Ulgen KO. Assessment of crosstalks between the Snf1 kinase complex and sphingolipid metabolism in S. cerevisiae via systems biology approaches. Mol Biosyst. 2013;9(11):2914-31. doi: 10.1039/c3mb70248k. PubMed PMID: 24056632.
- Borklu Yucel E, Ulgen KO. A network-based approach on elucidating the multi-faceted nature of chronological aging in S. cerevisiae. PLoS One. 2011;6(12):e29284. Epub 2012/01/05. doi: 10.1371/journal.pone.0029284PONE-D-11-14294 [pii]. PubMed PMID: 22216232; PubMed Central PMCID: PMC3244448.
- Hannun YA, Obeid LM. Many ceramides. J Biol Chem. 2011;286(32):27855-62. Epub 2011/06/23. doi: R111.254359 [pii]10.1074/jbc.R111.254359. PubMed PMID: 21693702; PubMed Central PMCID: PMC3151029.
- Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22(1):50-60. Epub 2011/10/18. doi: S0962-8924(11)00177-2 [pii]10.1016/j.tcb.2011.09.003. PubMed PMID: 22001186; PubMed Central PMCID: PMC3253987.