Production of Omega-3 and Omega-6 Polyunsaturated Fatty Acids by Metabolic Engineering of Yarrowia lipolytica
The Authors: Quinn Zhu and Ethel N. Jackson
Abstract
The omega-3 fatty acids, cis-5, 8,11,14,17-eicosapentaenoic acid (C20:5; EPA) and cis-4,7,10,13,16,19-docosahexaenoic acid (C22:6; DHA), have wide-ranging benefits in improving heart health, immune function, mental health, and infant cognitive development. Currently, the major source for EPA and DHA is from fish oil, and a minor source of DHA is from microalgae. To meet the increased demand for EPA and DHA, a clean and sustainable source of the omega-3 fatty acid EPA through fermentation was developed using metabolically engineered strains of Yarrowia lipolytica. The initial focus for commercial manufacture was for EPA production. EPA biosynthetic and supporting pathways were introduced into the oleaginous yeast to synthesize and accumulate high titers of EPA under fermentation conditions. This Yarrowia platform can also produce tailored omega-3 (EPA, DHA) and /or omega-6 (ARA, GLA) fatty acids mixtures in the cellular lipid profiles. Fundamental research such as metabolic engineering for strain construction, high through-put screening for strain selection, fermentation process development, and process scale-up were all needed to achieve the high levels of EPA titer, rate, and yield required for commercial application. Here we summarize how fundamental bioscience and industrial engineering combined to achieve large-scale production of Yarrowia biomass containing high amounts of EPA, which led to two commercial products.
The Need for a Sustainable Source of LCPUFAs
Omega-3 long chain polyunsaturated fatty acids (LCPUFAs), which include eicosapentaenoic acid (EPA, C20:5n-3) and docosahexaenoic acid (DHA, C22:6n-3), are natural products that are essential for human and animal health. EPA and DHA have crucial roles in the structure and function of cellular membranes and serve as precursors to several important eicosanoids including prostacyclins, leukotrienes and prostaglandins. There have been many clinical studies showing a wide range of health benefits from the omega-3 LCPUFAs, especially EPA and DHA (1-3). In general, it is believed that EPA is able to improve cardiovascular health, mental health, and immune function, while DHA is able to improve infant cognitive development, brain function, and eye health. The Japan EPA Lipid Intervention Study (JELIS) showed that EPA is a promising treatment for prevention of major coronary events (4). The AMR101 study also showed that pure EPA fatty acid significantly reduced triglyceride levels in adult patients with severe hypertriglyceridemia (5). The human body can only inefficiently synthesize EPA and DHA from omega-3 alpha-linolenic acid (C18:3; ALA), but cannot de novo synthesize them (2). EPA and DHA in our bodies are supplied largely from foods, especially cold-water marine fishes (3).
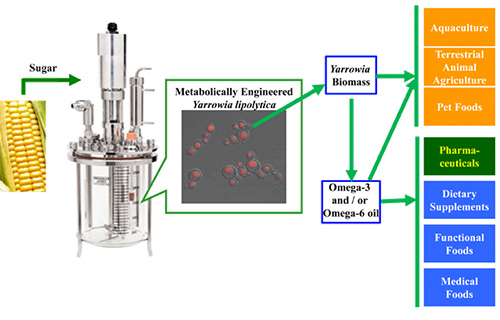
EPA and DHA are synthesized de novo in marine microorganisms and phytoplankton and accumulate in fish through the food chain. Some ocean fishes (e.g. Pacific sardine) can accumulate significant amounts of EPA and DHA by eating microalgae cells in the ocean. EPA and DHA are used in aquaculture, pharmaceuticals, human nutritional supplements, terrestrial animal feed, pet food and personal care. The demand for EPA and DHA is growing, but most commercially available EPA and DHA are produced using wild-caught ocean fish. Ocean fisheries are being depleted by over-fishing and are not a sustainable source for the increased demand. In addition, environmental contaminants, such as methylmercury, polychorinated biphenyls, dioxins, and several other halogenated, persistent organic pollutants (6), are being found in fish oils, creating further need for an alternative source of omega-3 fatty acids. A sustainable, land-based source is needed. To overcome this limitation, biotechnology industries started to produce DHA directly from microalgae in large-scale fermentation process (7). However, there was no large scale land-based EPA production from wild type organisms, because EPA productivity is too low to meet commercial targets. Consequently, a sustainable source of LCPUFAs has been developed by metabolic engineering of Yarrowia lipolytica (Fig. 1) to produce various omega-3 and omega-6 fatty acids. The first targeted product for commercialization was EPA due to its unique health benefits and the lack of a land-based sustainable supply. This metabolic engineering of Y. lipolytica now provides a commercially viable, sustainable, land-based source of EPA and other valuable LCPUFAs.
Figure 1. A sustainable source of LCPUFAs via metabolically engineered Yarrowia lipolytica (18, 22).
Here we summarize both metabolic engineering and fermentation process development for commercial production of EPA, including an engineered strain which produced EPA at more than 25% of its dry cell weight. The purified lipids from EPA producing strains have been used to develop a commercial product, NewharvestTM EPA oil, for a human nutritional supplement. The high-EPA biomass of these strains has also been used to raise Verlasso® (http://www.verlasso.com/) a sustainably farmed salmon. This technology paves the way for further improvement of EPA production strains, and development of strains with desired fatty acid compositions for specific applications.
Yarrowia lipolytica is a Safe and Productive Host for Omega-3 and Omega-6 Fatty Acids Production
Y. lipolytica is found primarily in foods with high proportions of fat and/or protein. Extensive research and analyses demonstrated that Y. lipolytica is a safe organism to be used for industrial applications (8). For example, it was classified as “Generally Recognized as Safe (GRAS)” for commercial production of food-grade citric acid (see the U.S. Food and Drug Administration list of microbial-derived ingredients approved for use in food: Title 21, Part 173, Sec. 165). It was also used to commercially produce a source of single-cell protein for animal feeds by British Petroleum (9). Y. lipolytica has an established history of robust fermentation performance. The cell density can reach more than 100 g DCW/L with carbohydrates such as glucose, fructose, glycerol or fatty acids as sole carbon source (10). Recently, Y. lipolytica strains have been engineered to use sucrose [11-12]. Most Y. lipolytica strains are haploid (13), but can also exist in diploid form. Depending on growth conditions, Y. lipolytica cells can differentiate into yeast, pseudomycelium and true mycelial forms (14-15). Y. lipolytica has a metabolism that is well suited to fatty acid production and lipid accumulation. Some Y. lipolytica strains are oleaginous organisms that can accumulate up to more than 30% DCW as storage triglycerides under the condition of nitrogen starvation and glucose excess. Although the central carbon metabolism of Y. lipolytica is similar to other yeasts, it has significant regulatory differences. It also has high flux for the pentose phosphate pathway that generates cofactor NADPH to support lipid biosynthesis (16-17). The lipid from glucose-grown cells is comprised mainly of triglycerides in which oleic acid (C18:1 n-9) and linoleic acid (LA, C18:2 n-6) are the two major fatty acids (10, 18).
There are six chromosomes in Y. lipolytica. A complete genome sequence of strain CLIB122 has been published (19). It has a total of about 20 Mb DNA that encodes about 6,500 genes. There is no extra-chromosomal plasmid discovered in wild-type strains. Genetic transformation occurs when exogenous DNA integrates into the genome by homologous and non-homologous recombination. Y. lipolytica has been used as a model system for studying hydrophobic substrate utilization, peroxisome biogenesis, lipid metabolism and bio-lipid production (20-21). It is easy to develop auxotrophic mutants for Y. lipolytica. Transformants can be selected by complementation of auxotrophic mutations and the use of antibiotic resistance genes as selectable markers is not required. The auxotrophic markers (10, 13, 22) most commonly used are the LYS5 genecoding for saccharopine dehydrogenase, the LEU2 gene coding forbeta-isopropylmalate dehydrogenase and the URA3 geneencoding for orotidine 5’-monophosphate decarboxylase. The counter selection system of the URA3 gene and 5-fluoroorotic acid (5-FOA) allows multiple rounds of integration of functional genes into the Y. lipolytica genome thereby to introduce many copies of foreign genes. In the last 30 years, Y. lipolytica has been one of the most studied non-conventional yeasts (13, 20). There is extensive knowledge accumulated on its genetics, molecular biology and physiology. Most of these studies suggest that Y. lipolytica is not only a good model system for basic scientific research, but also for industrial applications (18, 22).
We collected and screened over 40 different Y. lipolytica strains for their fermentation performances and ability to accumulate omega-3 fatty acids when these fatty acids are fed as substrates. The strain American Type Culture Collection (ATCC) #20362 achieved our fermentation performance targets: DCW greater than 100 g/L, lipid content greater than 30% DCW, and lipid productivity greater than 1 g/L/h. This ATCC #20362 strain was therefore chosen for pathway engineering. The genome sequence of ATCC #20362 has more than 99% identity with strain CLIB122 (www.genolevures.org/yali.html).
Biosynthesis Pathways to Produce Omega-3 and Omega-6 Fatty Acids
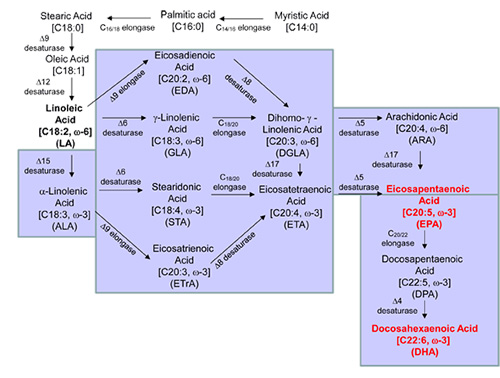
The wild-type strain ATCC #20362 (10, 18) can synthesize linoleic acid (LA, C18:2 n-6), but it cannot make omega-3 fatty acids. Different biosynthetic routes are known to make EPA and DHA, including the anaerobic polyketide synthase pathway (23) or an aerobic desaturase and elongase pathway (24). The microalgae, Crypthecodinium cohnii and Schizochytrium sp., used for DHA commercial production utilize the polyketide synthase pathway. Many microalgaeand some marine bacteria alsouse the polyketide synthase pathway to synthesize EPA (23, 25). However the EPA productivity of these organisms is insufficient for commercial production. The aerobic pathway (Fig. 2) can be further classified into a 6-desaturase pathway (the 6 pathway, found in algae, mosses, fungi and others) or a 9-elongase plus 8-desaturase (26) pathway (the 9 pathway). The 9 pathway (27) has been found in some species from Prymnesiophyceae (Pavlova, Isochrysis), Acanthamoebae (e.g. Acanthamoeba) and Euglenophyceae (e.g. Euglena). Genes from a variety of natural sources (10, 18, 22, 28-35) were introduced into Y. lipolytica to construct and optimize steps in the pathways outlined in Fig. 2.
Figure 2. Pathways for omega-3 EPA and DHA production (10, 24, 26, 27).
The engineered pathways for omega-3 EPA and DHA production are indicated in light blue.
The EPA and DHA are indicated in red.
Specifically, all pathways (Fig. 2) require the initial conversion of oleic acid to LA, the first of the omega-6 fatty acids, by a D12-desaturase (D12D). LA is used as substrate to produce EPA via D6 pathway as follows: (1) D6D converts LA to gamma-linolenic acid (GLA, 18:3n-6); (2) C18/20-elongase (C18E) converts GLA to dihomo-gamma-linoleic acid (DGLA, 20:3n-6); (3) D5-desaturase (D5D) converts DGLA to arachidonic acid (ARA, C20:4n-6), and (4): D17-desaturase (D17D) converts ARA to EPA. The “D6 pathway” can also use ALA as substrate to produce EPA: (1) D15-desaturase (D5D) converts LA to ALA; (2) D6D converts ALA to steridonic acid (STA, 18:4n-3); (3) C18E converts STA to ETA; and (4) D5D converts ETA to EPA.
LA is converted to EPA via the D9 pathway as follows: (1) D9 elongase (D9E) converts LA to eicosadienoic acid (EDA, 20:2n-6); (2) D8 desaturase (D8D) converts EDA to DGLA; (3) D5D converts DGLA to ARA; and (4) D17D convert ARA to EPA. The “D9 pathway” can also use ALA as a substrate to produce EPA as follows: (1) D15D converts LA to ALA; (2) D9E converts ALA to eicosatrienoic acid (ETrA: 20:3n-3); (3) D8D converts ETrA to ETA; and (4) D5D converts ETA to EPA.
It should be noted that both 15D and 17D are omega-3 desaturases; these two enzymes convert the omega-6 fatty acids into omega-3 fatty acids. The 15D converts LA to ALA. So far no 15D converts LA to ALA with 100% efficiency, so transformed cells with a heterologous 15D gene will contain both LA and ALA. Therefore, the 6 and 9 pathways can simultaneously use both LA and ALA as primary substrates. Apart from its primary function to convert ARA to EPA, most 17D can also convert EDA to ETrA with less efficiency.
For the synthesis of DHA from EPA, two additional steps are required: (1) C20/22-elongase (C20E) converts EPA to docosapentaenoic acid (DPA, 22:5n-3) and (2) delta4-desaturase (D4D) converts DPA to DHA. Some fish and seal oils contain omega-3 DPA. Incromega Trio oil, launched by Croda Ltd. in 2007, contains EPA (15%), DPA (7.5%), and DHA (30%). In some DHA oils originating from Schizochytrium (36), an omega-6 docosapentaenoic acid (DPA, 22:5n-6) is present that is not found in fish oil.
Metabolic Engineering of Y. lipolytica to Produce Omega-3 and Omega-6 Fatty Acids
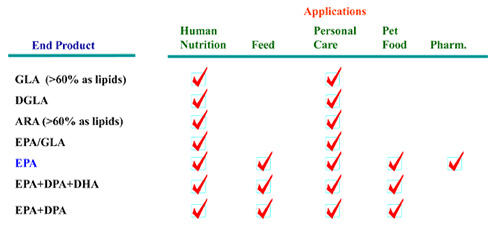
As shown in Figure 2, the omega-3 fatty acids ALA, STA, EPA, DPA, DHA, and the omega-6 fatty acids GLA, DGLA, ARA are all commercially relevant products which can be synthesized via these pathways. Y. lipolytica has been developed as a platform cell factory to produce all these omega-3 and omega-6 fatty acids (Table 1).
Table 1. Y. lipolytica metabolically engineered strains for production of omega-3 and omega-6 fatty acids
Co-expression of Δ12D and Δ6D in Y. lipolytica resulted in GLA accumulation at over 25% total lipids (28). Increasing the copy number of genes for of Δ12D and Δ6D, and over-expressing C16/C18 elongase (29) in a pex3 mutant, significantly increased GLA production to reach 60% total lipids (Wang, J; Bostick, M; and Zhu, Q; unpublished data). Similar strategies were employed to generate Yarrowia strains to produce DGLA and ARA to reach 42% and 35% total lipids, respectively (18). By increasing gene copy number for enzymes catalyzing ARA biosynthesis, smoothing the fatty acid trafficking in the ER, and other approaches, an ARA production strain has been generated to reach 60% of total lipids (Fan, X; Wilczek, JM; and Zhu, Q; unpublished data).
By using the D6 pathway, Y. lipolytica strain Y2097 was generated to produce lipids containing EPA at about 40% and GLA at about 21% (Fig. 2). The Y2097 strain contained nineteen copies of ten different heterologous genes that integrated into its genome and “pushed” 70% of total fatty acids into the engineered pathway (10).
By using the D9 pathway,Yarrowia strains have been generated to produce EPA as the first commercial product. A first generation strain Y4305 was developed to produce EPA at 56.6% of total lipids and about 15% of DCW (18). Strain Y4305 contains 30 copies of nine different heterologous genes. Additional strains (30) were later developed by a combination of several strategies (22). First, an efficient EPA biosynthetic pathway was built using strong promoters, codon-optimized heterologous genes, and multiple gene copies for each step. Second, the carbon flux was pushed toward the EPA biosynthesis pathway by over-expression of C16/C18 elongase (29) and D12D (31), and was pulled for EPA biosynthesis by using multiple copies of 17-desaturase genes [32]. Third, b-oxidation was eliminated by targeted deletion of peroxin genes(18, 33). Fourth, EPA trafficking was controlled by fine regulation of different acyltransferases (22, 30). The new commercial strain Z5567 containing 41 copies of 19 different genes produced EPA at 50% total lipids and 25% DCW. Two commercial products have been developed based on EPA production strains. The purified EPA lipids have been used to develop NewharvestTM EPA oil, for a human nutritional supplement. The high-EPA biomass has been used to raise Verlasso®, a sustainably farmed salmon.
Y. lipolytica strains to produce DHA have also been demonstrated (34). Based on strain Y4305, a DHA strain was constructed by over-expression of heterologous D4D and C20E genes, or expression of a multizyme having both D4D and C20E (35). The DHA lipid contains about 5% DHA, 13.5% DPA and 26.5% EPA, which is similar to the seal oil (37) that contains high amounts of beneficial omega-3 DPA (38). It is obvious that Y. lipolytica strains based on our improved EPA production strains can be generated to produce much higher amounts of DHA. These studies have demonstrated that Y. lipolytica is a good host for metabolic engineering for production of omega-3 and omega-6 fatty acids. Strains can be developed to produce cost-effective products with desired fatty acid compositions for specific applications.
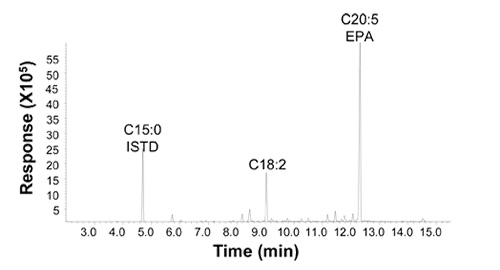
The fatty acid profile of the lipids in selected EPA strains shows that the EPA content is more than 55% of the total lipid, and the saturated fatty acid, C16:0 plus C18:0, is less than 5%. The only major intermediate was LA, at about 17% (Fig. 3). Lipid distribution data show that more than 85% of the fatty acids were in triglycerides form, and the 55% of the EPA is at the sn-1 and sn-3 positions and only 35% at the sn-2 position of the glycerol backbone.
Figure 3. Fatty acid profile of EPA lipids produced by metabolically engineered strain Y4305 (18). The fatty acid composition was determined by gas chromatography analyses
The saturated fatty acids in the EPA lipid produced by strain Y4305 is extremely low (18). This is due to both the characteristics of the D9 pathway and the carefully balanced expression of different genes in the pathway. In engineered Y. lipolytica strains, the substrate conversion efficiency [calculated as product/(product + substrate) × 100%] of the introduced desaturases is significantly higher than the elongases. This is the result of the differences in substrate and availability. Desaturation occurs on the acyl moiety of phospholipid (PL), elongation occurs on the acyl moiety of the acyl-CoAs, and the acyl-CoA pool is limited, leading to lower conversion efficiency by elongases (18). The selection of the D-9 pathway ensured that the rate limiting elongation is the first step of the engineered pathway. Accumulation of intermediates is therefore kept to a minimum, in contrast to cells engineered with the D-6 pathway where the first step is not rate limiting and accumulation of GLA becomes significant (10).
The EPA lipid produced by this metabolically engineered Y. lipolytica is very healthy and unique. Likewise, other omega-3 and omega-6 fatty acids with desired composition can also be produced from this technology platform. This land-based, sustainable production of vegetarian omega-3 and omega-6 lipids is a superior source for these essential molecules for applications in nutritional supplements, functional foods, infant foods, medical foods, pharmaceuticals, and animal feeds.
Laboratory-Scale and Commercial-Scale Fermentations for EPA Production
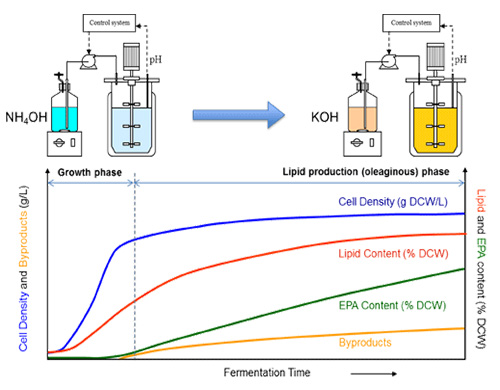
Fermentation process development is a necessary step for both strain construction and commercial manufacturing of omega-3 and omega-6 fatty acids. Fermentation research was carried out in parallel with strain engineering (22). Fermentation experimentation included (1) strain screening under fermentation conditions, (2) optimization of fermentation conditions for promising new strains, and (3) process scale-up. After top candidate strains were selected by micro-fermentation (39) and laboratory-scale fermentation analyses, fermentation optimization work was conducted (22). Since the EPA lipid is an intracellular product of the Yarrowia biomass, the goal of optimization is first to maximize the biomass production in growth phase, and then to maximize EPA production and minimize byproduct formation in the oleaginous phase. A two-stage fed-batch fermentation process was developed to maximize both biomass and EPA production (Figure 4). In the first stage of the fermentation, the Yarrowia cells are grown on the carbohydrate substrate with excess nitrogen. In the second stage, the nitrogen supply is limited, while the carbohydrate feed continues. Cell growth then stops after consuming the residual nitrogen in the medium and the Yarrowia cells start to accumulate lipids. Optimal conditions were often found to vary when the strain was engineered in different genetic backgrounds. Therefore, more optimization work is expected to improve a newly selected strain’s performance.
Figure 4. A two-stage fermentation process for omega-3 EPA production (22).
In growth phase, nitrogen is mainly provided by NH4OH for pH control to build up biomass.
During the production phase, nitrogen is limited by switching the ammonium base to KOH to produce lipid with high EPA content.
A critical aspect of process development and scale-up for the EPA project was the use of dynamic models (40-42). Unstructured mathematical models were built from first principles, which included the model equations of cell growth, substrate consumption, nitrogen utilization, oxygen uptake, lipid and EPA formation, and byproduct accumulation. The models were matched to the historical experimental data from many lab-scale and pilot-scale fermentation experiments under different conditions. The models could predict cell growth, DCW, dissolved oxygen level, oxygen uptake rate, CO2 evolution rate, and other variables that were also measured during the fermentation as a function of various medium and process conditions. The dynamic models were thus able to predict the key performance parameters (e.g. titer, rate, and yield of a product) before and during the run, and further help guide the fermentation optimization and process scale-up (22).
The last step in fermentation process development is the scale-up to pilot and then commercial scales. There are a few criteria commonly used for fermentation scale-up, including geometry similarity, power input, mass transfer coefficient KLa (40). For the EPA fermentation process, the selected production strain was tested in the pilot-scale facilities before setting criteria for scale up, in order to provide information about the pilot-scale fermentation’s dynamic behavior. Besides the regular online data of temperature, pH value, feed rate, and dissolved oxygen levels, the dynamic information also included the agitation, aeration rate, and the mass transfer characteristics. The benefits of highly predictive dynamic models became even more evident as the process moved from the pilot plant to commercial production. Data from the pilot-scale fermentations were incorporated into the dynamic models, which then were used to predict the commercial-scale fermentation performance in the commercial-scale fermenters These models plus “scale-down” experiments guided the successful scale-up (22).
Summary
The yeast, Yarrowia lipolytica, was engineered as a cell factory platform to produce both omega-3 and omega-6 fatty acids (Table 1). By over-expressing a combination of enzymes that are necessary for synthesis of EPA via the 9/8 pathway and for optimization of lipid metabolism, a strain was created that is capable of making 25% EPA as dry biomass and more than 50% lipids as EPA. The high level of EPA production was achieved through careful balancing of the expression levels of various pathway enzymes, and modification of fatty acid and lipid metabolism of the host. Disruption of the peroxisome biogenesis gene had a major positive impact on the production of EPA and the metabolism of storage lipid, as well as reduction of the major by-products. Two commercial products, NewharvestTM EPA oil and Verlasso® salmon were developed using our sustainable EPA source. These studies have demonstrated that Y. lipolytica is an advantageous organism for metabolic engineering for production of omega-3 and omega-6 fatty acids. Strains can be developed to produce cost-effective products with desired fatty acid compositions for specific applications. These results exemplify the way in which biotechnology can generate novel products and bring to market alternatives that can replace those from non-sustainable traditional sources. This use of metabolic engineering to produce a key chemical currently sourced from a non-sustainable resource is an example of the power of biotechnology in the push towards a sustainable society (43).
References:
- Chacon-Lee, T.L. and Gonzalez-Marino, G.E. Microalgae for “Healthy” Foods-Possibilities and Challenges. Comprehensive Reviews in Food Science and Food Safety, 9, 655–675 (2010) (doi:10.1111/j.15414337.2010.00132.x).
- Kapoor, R. and Patil, U.K. Importance and production of omega-3 fatty acids from natural sources. International Food Research Journal, 18, 493-499 (2011).
- Martins, D.A., Custódio, L., Barreira, L., Pereira, H., Ben-Hamadou, R., Varela, J., and Abu-Salah, K.M. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs, 11, 2259-2281 (2013) (doi:10.3390/md11072259).
- Yokoyama, M., Origasa, H., Matsuzaki, M., Matsuzawa, Y., Saito, Y., Ishikawa, Y., Oikawa, S., Sasaki, J., Hishida, H., Itakura, H., Kita, T., Kitabatake, A., Nakaya, N., Sakata, T., Shimada, K., Shirato, K. Effects of eicosapentaenoic acid on major coronary events in hypercholesterolaemic patients (JELIS): A randomised open-label, blinded endpoint analysis. The Lancet, 369, 1090-1098 (2007).
- Ballantyne, C.M., Bays, H.E., Kastelein, J.J., Stein, E., Isaacsohn, J.L., Braeckman, R.A., Soni, P.N. Efficacy and safety of eicosapentaenoic acid ethyl ester (AMR101) therapy in statin-treated patients with persistent high triglycerides (from the ANCHOR study). Am. J. Cardiol. 110, 984-992 (2012) (doi: 10.1016/j.amjcard.2012.05.031).
- Costa, L.G. Contaminants in fish: risk-benefit considerations. Arh. Hig. Rada. Toksikol. 58, 567-74 (2007).
- Kyle, D.J. The large-scale production and use of single-cell highly enriched oil in docosahexaenoic acid. In: Shahidi F, Finley J (Eds) Omega-3 Fatty Acids: Chemistry, Nutrition, and Health Effects. Oxford University Press, pp 92-107 (2001).
- Groenewald, M., Boekhout, T., Neuvéglise, C., Gaillardin, C., van Dijck, P.W., Wyss, M. Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 40,187-206 (2014) (doi: 10.3109/1040841X).
- Ratledge, C.; Single cell oils for the 21st century. Single Cell Oils. Cohen, Z.; Ratledge, C. Eds.; AOCS Press: Champaign, Illinois, 1-20 (2005).
- Zhu, Q., Xue, Z., Yadav, N., Damude, H., Pollak, D., Rupert, R., Seip, J., Hollerbach, D., Macool, D., Zhang, H. Metabolic engineering of an oleaginous yeast for the production of omega-3 fatty acids. In: Cohen Z and Ratledge C (Eds) Single Cell Oils, 2nd Ed. AOCS Press, Urbana Illinois. pp51-73 (2010).
- Hong, S-Y., Seip, J., Walters-Pollak, D., Rupert, R., Jackson, R., Xue, Z., Zhu, Q. Engineering Yarrowia lipolytica to express secretory invertase with strong FBA1IN promoter. Yeast, 29, 59–72(2011) (DOI: 10.1002/yea.1917).
- Lazar, Z., Rossignol, T., Verbeke, J., Crutz-Le Coq, A.M., Nicaud, J.M., Robak, M. Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. J. Ind. Microbiol. Biotechnol. 40, 1273-1283 (2013) (doi: 10.1007/s10295-013-1323-1).
- Barth, G., Gaillardin, C. Yarrowia lipolytica. In: Wolf K (Ed) Nonconventional Yeasts in Biotechnology. Springer-Verlag, Berlin, Heidelberg, New York, pp313-388 (1996).
- Pérez-Campo, F.M., Domínguez, A. Factors affecting the morphogenetic switch in Yarrowia lipolytica. Curr. Microbiol. 43, 429-433 (2001).
- Szabo, R., Stofaníková, V. Presence of organic sources of nitrogen is critical for filament formation and pH-dependent morphogenesis in Yarrowia lipolytica. FEMS Microbiol. Lett. 206, 45-50 (2002).
- Blank, L.M., Lehmbeck, F., Sauer, U. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 5, 545-558 (2005).
- Christen, S., Sauer, U. Intracellular characterization of aerobic glucose metabolism in seven yeast species by 13C flux analysis and metabolomics. FEMS Yeast Res. 11, 263-272. (2011) (doi: 10.1111/j.1567-1364.2010.00713.x.).
- Xue, Z., Sharpe, P.L., Hong, S.P., Yadav, N.S., Xie, D., Short, D.R., Damude, H.G., Rupert, R.A., Seip, J.E., Wang, J., Pollak, D.W., Bostick, M.W., Bosak, M.D., Macool, D.J., Hollerbach, D.H., Zhang, H., Arcilla, D.M., Bledsoe, S.A., Croker, K., McCord, E.F., Tyreus, B.D., Jackson, E.N., and Zhu, Q. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nature Biotech. 31, 734-740 (2013) (doi:10.1038/nbt.2622).
- Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., Lafontaine, I., Montigny, J., Marck, C., Neuvéglise, C., Talla, E., Goffard, N., Frangeul, L., Aigle, M., Anthouard, V., Babour, A., Barbe, V., Barnay, S., Blanchin, S., Beckerich, J.M., Beyne, E., Bleykasten, C., Boisramé, A., Boyer, J., Cattolico, L., Confanioleri, F., Daruvar, A., Despons, L., Fabre, E., Fairhead, C., Ferry-Dumazet, H., Groppi, A., Hantraye, F., Hennequin, C., Jauniaux, N., Joyet, P., Kachouri, R., Kerrest, A., Koszul, R., Lemaire, M., Lesur, I., Ma, L., Muller, H., Nicaud, J.M., Nikolsk, M., Oztas, S., Ozier-Kalogeropoulos, O., Pellenz, S., Potier, S., Richard, G.F., Straub, M.L., Suleau, A., Swennen, D., Tekaia, F., Wésolowski-Louvel, M., Westhof, E., Wirth, B., Zeniou-Meyer, M., Zivanovic, I., Bolotin-Fukuhara, M., Thierry, A., Bouchier, C., Caudron, B., Scarpelli, C., Gaillardin, C., Weissenbach, J., Wincker, P., Souciet, J.L. Genome evolution in yeasts. Nature, 430,35-44 (2004).
- Nicaud, J.M. Yarrowia lipolytica. Yeast, 29, 409–418 (2012) (DOI: 10.1002/yea.2921).
- Tai, M., Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15, 1-9 (2013) (doi: 10.1016/j.ymben.2012.08.007).
- Xie, D., Jackson, E.N. and Zhu, Q. Sustainable Source of Omega-3 Eicosapentaenoic acid from Metabolically Engineered Yarrowia lipolytica: From Fundamental Research to Commercial Production. Appl. Microbiol. Biotechnol. 99, 1599-1610 (2015) (doi: 10.1007/s00253-014-6318-y).
- Metz, J.G., Roessler, P., Facciotti, D., Levering, C., Dittrich, F., Lassner, M., Valentine, R., Lardizabal, K., Domergue, F., Yamada, A., Yazawa, K., Knauf, V., Browse, J. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science, 293. 290-293 (2001).
- Meesapyodsuk, D., Qiu, X. The front-end desaturase: structure, function, Evolution and biotechnological use. Lipids, 47, 227-237 (2012) (doi: 10.1007/s11745-011-3617-2).
- Wen, Z., Chen, .F. Prospects for eicosapentaenoic acid production using microorganisms. In: Cohen Z and Ratledge C (Eds) Single Cell Oils. AOCS Press, Champaign, Illinois, pp138-160 (2005).
- Wallis, J.G., Browse, J. The Delta8-desaturase of Euglena gracilis: an alternate pathway for synthesis of 20-carbon polyunsaturated fatty acids. Arch. Biochem. Biophys. 365, 307-316 (1999).
- Sayanova, O., Napier, J.A. Eicosapentaenoic acid: biosynthetic routes and the potential for synthesis in transgenic plants. Phytochemical, 65,147-158 (2004).
- Zhu, Q., Walters-Pollak, D.M. Delta 12 and Delta 6 desaturases able to catalyze the conversion of oleic acid to GLA have been introduced into the genome of Yarrowia, using zeta-directed integration. US Patent, US7465564 (2008).
- Macool, D.J., Xue, Z., Zhu, Q. A Mortierella alpina C16/18 fatty acid elongase. US Patent, US7470532 (2008).
- Hong, S-Y, Sharpe, P.L., Xue, Z., Yadav, N.S., Zhang, H., Zhu, Q. Recombinant microbial host cells for high eicosapentaenoic acid production. US Patent, US8703473 (2014).
- Yadav, N.S., Zhu, Q., Zhang, H. 12 desaturases suitable for altering levels of polyunsaturated fatty acids in oleaginous yeast. US Patent, US7504259 (2009).
- Xue, Z., He, H., Hollerbach, D., Macool, D.J., Yadav, N.S., Zhang, H., Szostek, B., Zhu, Q. Identification and characterization of new Δ-17 fatty acid desaturases. Appl. Microbiol. Biotechnol. 97, 1973-1985 (2013) (doi: 10.1007/s00253-012-4068-2).
- Hong, S.P., Sharpe, P..L, Xue, Z., Yadav, N.S., Zhu, Q.Q. Peroxisome biogenesis factor protein (pex) disruptions for altering polyunsaturated fatty acids and total lipid content in oleaginous eukaryotic organisms. US Patent Application 20090117253 (2009).
- Damude, H.G., Macool, D.J., Picataggio, S.K., Ragghianti, J.J., Seip, J.E., Xue, Z., Yadav, N.S., Zhang, H., Zhu, Q. Docosahexaenoic acid producing strains of Yarrowia lipolytica. US Patent, US7550286 (2009).
- Damude, H.G., Kinney, A.J., Ripp, K.G., Zhu, Q. Multizymes and their use in making polyunsaturated fatty acids. US Patent Application, US20080254191 (2008).
- Barclay, W.C. Weaver, Metz, J. Development of a docosahexaenoic acid production technology using Schizochytrium: a historical perspective. Single Cell Oils. Cohen, Z.; Ratledge, C. Eds.; AOCS Press: Champaign, Illinois, pp36-52 (2005).
- Shahidi, F., Wanasundara, P.K.J. and Wanasundara, U.N. Seal blubber oil: A novel source of w3 fatty acids. J. Food Lipids, 3, 293-306 (1996).
- Kaur, G., Cameron-Smith, D., Garg, M., Sinclair, A.J. Docosapentaenoic acid (22:5n-3): A review of its biological effects. Prog. Lipid Res. 50, 28-34 (2011) (doi: 10.1016/j.plipres.2010.07.004).
- Xie, D. Using an advanced microfermentor system for strain screening and fermentation optimization. In: Cheng Q (Ed) Methods in Molecular Biology, 834 (Microbial Metabolic Engineering), Humana Press, New York. pp217-231 (2012).
- Shuler, M.L., Kargi, F. Biorocess Engineering, 2nd edition. Prentice Hall PTR, Upper Saddle River, NJ, pp286-306 (2002).
- Ozbek, B. Mathematical modeling and simulation studies of scale-down fermentation systems. Chem. Eng. Technol. 20, 259-267 (1997).
- Nienow, A.W., Nordkvist, M., Boulton, C.A. Scale-down/scale-up studies leading to improved commercial beer fermentation. Biotechnol. J. 6, 911-925 (2011) (doi: 10.1002/biot.201000414).
- Wynn, J.P. Taking fish out of fish oil. Nature Biotech. 31, 716-717 (2013) (doi: 10.1038/nbt.2656).