Biosynthesis of Fatty Acids
The Author: Dr. Peter Reilly, Department of Chemical and Biological Engineering, Iowa State University, Ames, Iowa 50011, U.S.A.
Reactions of the fatty acid synthesis cycle
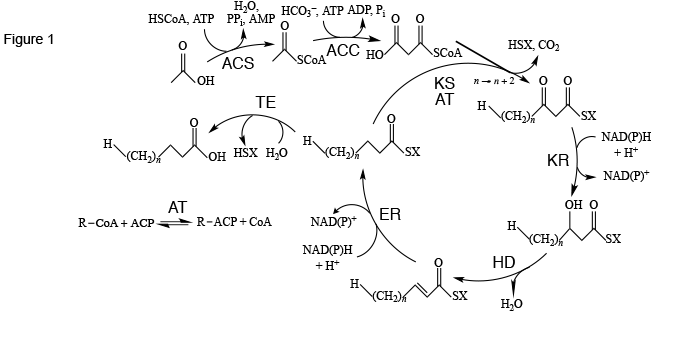
The standard way for cells to synthesize fatty acids is through the fatty acid synthesis cycle (Figure 1). This cycle of eight enzymes (acyl-CoA synthase, acyl-CoA carboxylase, acyltransferase, ketoacyl synthase, ketoacyl reductase, hydroxyacyl dehydratase, enoyl reductase, and thioesterase) and acyl carrier protein) is initated with acetic acid, CoA, and ATP to make acetyl-CoA using acyl-CoA synthase as catalyst. A second step, using another ATP and bicarbonate ion catalyzed by acyl-CoA carboxylase, yields malonyl-CoA. With ketoacyl synthase and acyltransferase catalysis, malonyl-CoA is added to an acyl chain, usually activated with acyl carrier protein, to make an acyl chain two methylene groups longer. Further reduction, dehydration, and reduction with ketoacyl synthase, hydroxyacyl dehydratase, and enoyl reductase catalysis, respectively, leads to a saturated and unhydroxylated acyl chain activated with acyl carrier protein. If the chain is of appropriate length, it is attacked by thioesterase to release acyl carrier protein, yielding the finished fatty acid.
Figure 1. The fatty acid synthesis cycle and the enzyme groups that are part of it. ACC: acetyl-CoA carboxylase; ACS: acyl-CoA synthase; AT: acyltransferase; ER: enoyl reductase; HD: hydroxyacyl dehydratase; KR: ketoacyl reductase; KS: ketoacyl synthase; TE: thioesterase. SX: Coenzyme A or acyl carrier protein. Reproduced in modified form with permission from ref. 1. Copyright 2011, Oxford University Press.
Definitions of enzyme families and clans
It is perhaps not fully appreciated that these eight enzyme groups and one carrier protein are, in fact, usually assemblages within each group of several (or many) different proteins not necessarily related to each other by amino acid sequence (primary structure) or even by three-dimensional structure (tertiary structure). Instead, unrelated proteins can be shaped by convergent evolution to catalyze the same general reaction. Conversely, proteins of related primary and tertiary structures produced by different organisms but catalyzing the same reaction can be part of families whose members are generally considered to have the same common ancestor. Members of these families can be identified by automated interrogation of databases holding enzyme primary structures, where family members must pass a probability test of similarity followed by further multiple sequence alignment and comparison of tertiary structures (if available). In a further step, different families having unrelated (or only slightly related) primary structures but similar tertiary structures, and catalyzing the same general reaction by the same mechanism, can be gathered into clans within the same group. This is confirmed by computationally overlapping the tertiary structures of members of different families to confirm the low root mean square deviations of adjacent amino acid residues. Members of different families in the same clan are considered to have more distant common ancestors than those in the same family. A third diversification, not to be covered here, is that of family members into subfamilies, governed by statistical formulations applied to primary structures.
The ThYme database
The families of the enzymes catalyzing the reactions leading to fatty acid synthesis have been tabulated in the ThYme (Thioester-active enzYme) database [1]. At present this database consists of five families of acyl-CoA synthases, four families of acyl-CoA carboxylase subgroups (one of biotin carboxylases, one of biotin carboxyl carrier proteins, and two of carboxyl transferases) [2], one of acyltransferases, five of ketoacyl synthases [3], four of ketoacyl reductases [4], eight of hydroxyacyl dehydratases [4], five of enoyl reductases [4], 25 of thioesterases [5], and 16 of acyl carrier proteins [6] (Table 1). Although not all enzyme groups have been analyzed, one clan of ketoacyl synthases encompasses four of its five families [3], one clan of ketoacyl reductases covers three of its four families [7], one clan of hydroxyacyl dehydratases takes in two of its eight families [7], and four clans of thioesterases cover four, three, three, and two of its 25 families [5].
Most ThYme entries have resulted from genomic studies and have not been subjected to experimental projects. Therefore they usually do not have firmly established titles, often having been characterized as “uncharacterized proteins”, “hypothetical proteins”, or by other indefinite or even clearly incorrect terms. Databases such as ThYme have been helpful, by their use of primary structures to classify proteins, in directing these less characterized proteins into families containing other more studied and clearly described proteins.
Let us now more carefully consider each fatty acid synthesis cycle enzyme group.
Acyl-CoA synthases
Acyl-CoA synthases catalyze the first step in the fatty acid synthesis pathway, activating acetic acid with CoA and with the expenditure of an ATP molecule, leading to the formation of AMP, pyrophosphate, and water. (Figure 1). They are sorted into five families (Table 1), some with many more primary structures than others. Members of three families have similar tertiary structures (folds), while two have yet no known folds. As is common over the different groups of the cycle, the different families have different principal names and different Enzyme Commission (EC) numbers [8]. In some large families, as in family ACS1 here, there are so many different perceived enzyme functions that no single EC number can be assigned to the family. This divergence of families sorted by primary structure similarity, on one hand, and EC numbers based on enzyme substrates and products and therefore on the reactions they catalyze, on the other hand, further illustrates the phenomenon of convergent evolution of protein structures to serve the same purpose.
Archaea produce members of four of the five families, bacteria all five, and eukaryota two of the five (with a few members in a third family). Most eukaryotic members are produced by some combination of fungi, animals, and plants, as well as sometimes by algae and occasionally by other simple life forms.
Acyl-CoA carboxylases
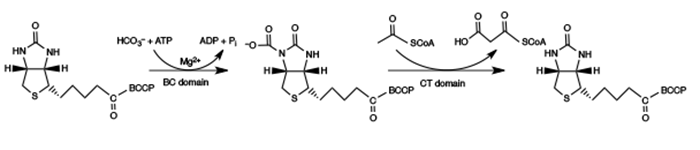
The reaction catalyzed by acyl-CoA carboxylase is superficially a simple one: acetyl-CoA being carboxylated with CO2 (HCO3– under physiological conditions) and ATP to form malonyl-CoA, with ADP and inorganic phosphate being liberated (Figure 1). However, the reaction is actually much more complex, with biotin attached to a biotin carboxyl carrier protein being carboxylated by a biotin carboxylase domain. The carboxyl group is then transferred to an acetyl-CoA molecule, catalyzed by a carboxyl transferase domain (Figure 2). The whole complex of biotin carboxylase, biotin carboxyl carrier protein, and carboxyl transferase makes up acyl-CoA carboxylase. A recent and very complete review of this enzymatic system has been published by Lombard and Moreira [9].
Figure 2. Schematic of an acetyl-CoA carboxylase-catalyzed reaction producing malonyl-CoA. Reproduced with permission from ref. 2. Copyright 2012, Springer Science+Business Media B.V.
A comparison of primary structures shows that all biotin carboxylases and biotin carboxyl carrier proteins make up two single families, BC1 and BCCP1, while carboxyl transferases are part of two families, CT1 and CT2 [2] (Table 1).
The placement of the biotin carboxylase, biotin carboxyl carrier protein, and carboxyl transferase domains is very complex. In most eukaryotes, acetyl-CoA carboxylases occur as biotin carboxylase-biotin carboxyl carrier protein-fused carboxyl transferase chains. However, in bacterial and eukaryotic acyl-CoA carboxylases more active on propionyl-CoA, 3-methylcrotonyl-CoA, and geranyl-CoA, the biotin carboxyl carrier protein and biotin carboxyl domains are attached to each other, but they are separate from the fused carboxyl transferase domains. In archaeal biotin-dependent carboxylases specific for acetyl-CoA and propionyl-CoA, biotin carboxyl carrier protein is separate from the biotin carboxylase and fused carboxyl transferase domains [9].
Acyltransferases
The acyltransferases are made up of one very large family (Table 1) whose members are produced by bacteria, eukaryota, and a few archaea. The specific task of the acyltransferases is to exchange the CoA moiety on the malonyl-CoA entering the fatty acid synthesis cycle with an acyl carrier protein (Figure 1).
Ketoacyl synthases
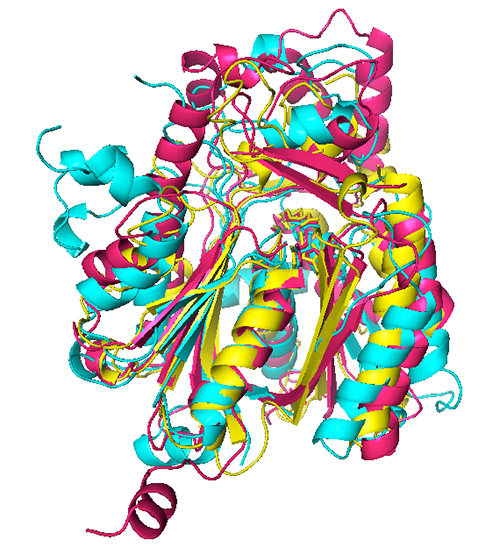
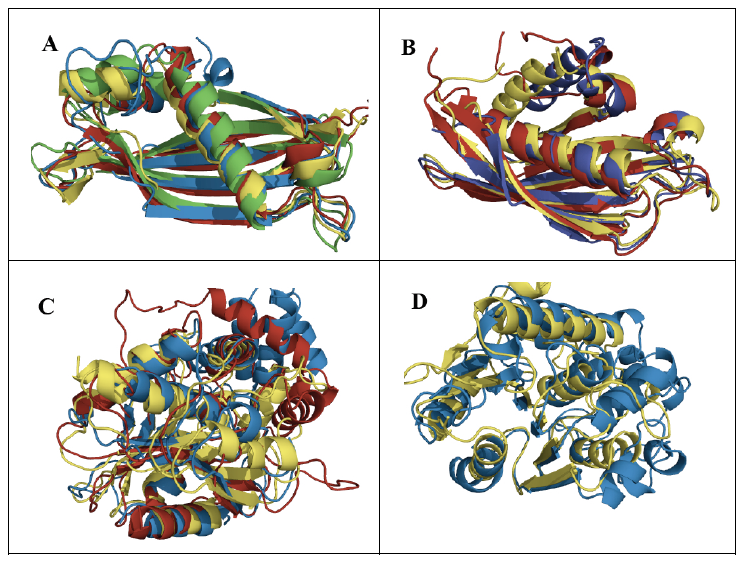
The general role of ketoacyl synthases, along with acyltransferases, is to add malonyl-CoA to an acyl-acyl carrier protein chain, with the loss of carbon dioxide and CoA and with the acyl chain increasing in length by two methylene groups [3] (Figure 1). There are five families of ketoacyl synthases, three of whose members have similar tertiary structures (Figure 3). Together with the members of a fourth family somewhat related to them by primary structure and by catalytic mechanism (members of all four families have a catalytic triad of Cys, His, and Asn or His), they form a single clan [3] (Table 1). These four families have very different functions, the members of one of them being involved in chalcone and stilbene synthesis. The fifth family, without a known tertiary structure, is made up only by eukaryotic enzymes involved in the elongation of already very long fatty acid chains.
Figure 3. Superimposed ketoacyl synthase (KS) crystal structures. KS1 Escherichia coli β-ketoacyl-ACP synthase III (yellow), KS3 Saccharopolyspora erythraea DEBS 2 (cyan), and KS4 Arachis hypogaea stilbene synthase (pink). Reproduced with permission from ref. 3. Copyright 2011, The Protein Society.
The β-oxidation pathway
Three enzymes, ketoacyl reductase, hydroxyacyl dehydratase, and enoyl reductase, are required to remove the 3-keto group added to the growing acyl chain by ketoacyl synthase (Figure 1). All three enzymes are also involved in the β-oxidation pathway, where fatty acids are broken down instead of being built up. In this pathway the ketoacyl reductase-, hydroxyacyl dehydratase-, and enoyl reductase-catalyzed steps operate in reverse, so these enzymes are given names characteristic of the reverse reactions. Oxidation of acyl-CoA catalyzed by acyl-CoA dehydrogenase (equivalent to enoyl reductase) with a flavin adenine dinucleotide (FAD) prosthetic group gives 2-enoyl-CoA. Hydration of 2-enoyl-CoA yielding 3-hydroxyacyl-CoA is catalyzed by 2-enoyl hydratase (equivalent to hydroxyacyl dehydratase). The third step, oxidation of 3-hydroxyacyl-CoA to give 3-ketoacyl-CoA by catalysis with L-3-hydroxyacyl-CoA dehydrogenase (equivalent to ketoacyl reductase) using nicotinamide adenine dinucleotide (NAD+). A final step, with addition of CoA, removes an acetyl-CoA molecule and leaves an acyl-CoA molecule two carbon atoms shorter than before. It is catalyzed by 3-ketoacyl-CoA thiolase, which is not a member of the standard fatty acid synthesis cycle.
Polyketide synthesis
A short paragraph can serve to describe the difference between fatty acid synthesis and polyketide synthesis. In the latter, the ketoacyl reductase-, hydroxyacyl dehydratase-, and enoyl reductase-catalyzed steps do not necessarily occur during each turn around the cycle, so keto and hydroxy groups and double bonds may remain in the lengthening chain. Furthermore, the chain can be elongated by substances other than malonyl-CoA, therefore leading to chains with odd numbers of carbon atoms or different functional groups. Finally, if chain growth is terminated not by water but by a hydroxy group, chain cyclization can occur.
Ketoacyl reductases
The ketoacyl reductases are divided into four families (Table 1), with family KR1 having the largest number of tabulated primary structures in the ThYme database (Table 1). This family includes many reductases and dehydrogenases that do not catalyze reactions associated with the fatty acid synthesis or polyketide synthesis cycles or the β-oxidation pathway [4]. It also includes the former family ER1, as all the amino acid sequences of these enoyl reductases were found also in family KR1. Members of families KR1, KR3, and KR4 have NAD(P)-binding Rossmann folds and serine, tyrosine, and lysine catalytic residues, and are part of clan KR-A [4,7]. Those in family KR2 have histidine and glutamate catalytic residues, but also NAD(P)-binding Rossmann folds along with C-terminal 6-phosphogluconate dehydrogenase folds. The Rossmann fold is composed of up to seven mainly parallel β-strands, with an α-helix between the first two β-strands. Members of families KR1 and KR2 are freestanding, while those of families KR3 and KR4 are parts of multi-enzyme chains carrying out fatty acid and polyketide synthesis.
Hydroxyacyl dehydratases
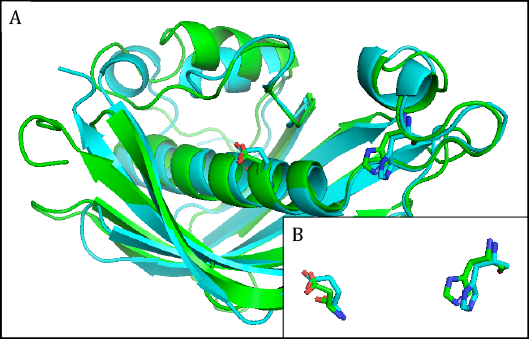
The hydroxyacyl dehydratases are found in eight families, of which families HD1 and HD3 through to HD6 have members with HotDog tertiary structures, while members of family HD2 have ClpP/crotonase folds [4] (Table 1). The HotDog fold is composed of one or occasionally two α-helices encased in a β-sheet made up of several β-strands, a good example being shown in Figure 4. No tertiary structures are yet known for members of families HD7 and HD8. Although tertiary structures of members of families HD1, HD5, and HD6 can be closely superimposed on each other, the catalytic aspartate and histidine residues of HD1 are not in the same location as the catalytic aspartate and histidine residues of HD5 or the glutamate and histidine residues of HD6 [7]. The latter pairs of residues, on the other hand, overlap each other very closely and therefore allow HD5 and HD6 to make up clan HD-A (Figure 4). Separately, HD3 and HD4 members are part of multi-enzyme fatty acid and polyketide synthases.
Figure 4. A) Superimposed representative hydroxyacyl dehydratase (HD) tertiary structures of clan HD-A: HD5 β-hydroxydecanoyl thiol ester-ACP dehydrase from Escherichia coli (green) and HD6 ((3R)-hydroxyacyl-ACP dehydratase (FabZ) from Helicobacter pylori (cyan). B) Superimposed active-site side chains of the same HD family representatives, with colors as in A. Reproduced with permission from ref. 7. Copyright 2014, Springer Science+Business Media Dordrecht.
Enoyl reductases
As previously noted, family ER1 has been merged into family KR1 because all of its entries are also found there. Furthermore, all members of each family catalyze the same types of reductive reactions. The remaining enoyl reductases fall into five families, of which those in families ER3 and ER4 are part of larger fatty acid and polyketide synthase complexes [4] (Table 1). Families ER2 and ER4 have NAD(P)-binding Rossmann folds, while members of family ER5 have GrosES-like structures, and those of family ER6 have TIM (β,α)-barrel folds.
A superimposition of a ER2 tertiary structure upon the structures of KR1, KR3, and KR4, members of clan KR-A, is extremely close, suggesting that ER2 members and members of the three ketoacyl reductase families have a distant common ancestor [7].
Thioesterases
The thioesterases are the most varied of the eight enzyme groups making up the fatty acid synthesis cycle. They are arranged into 25 families [5] (Table 1), most of which have members with either α/β-hydrolase or HotDog tertiary structures (Figure 5). Twelve of these families can be gathered into clans. Families TE5, TE9, TE10, and TE12 with HotDog folds are part of clan TE-A, while families TE8, TE11, and TE13 with different HotDog tertiary structures make up clan TE-B. Meanwhile families TE16, TE17, and TE18 with α/β-hydrolase folds are part of clan TE-C, and families TE20 and TE21 with different α/β-hydrolase tertiary structures make up clan TE-D. The mechanism of the thioesterases with α/β-hydrolase tertiary structures is generally accepted to be based on the Ser/His/Asp catalytic triads usually found with such folds. The mechanisms of those thioesterases with HotDog tertiary structures are much less known [5]. They may well have different mechanisms from each other, as their experimentally identified catalytic residues vary.
Figure 5. Superimposed representative thioesterase (TE) tertiary structures of A) clan TE-A: acyl-CoA hydrolases from TE5 Escherichia coli (green), TE9 Helicobacter pylori (red), TE10 Pseudomonas sp. (yellow), and TE12 Prochlorococcus marinus (blue); B) clan TE-B: acyl-CoA hydrolases from TE8 Homo sapiens (blue), TE11 Arthrobacter sp. (red), and TE13 Escherichia coli (yellow); C) clan TE-C: acyl-ACP hydrolases from TE16 Homo sapiens (blue), TE17 Saccharopolyspora erythraea (red), and TE18 Amycolatopsis mediterranei (yellow); D) clan TE-D: protein-acyl hydrolases from TE20 Bos taurus (blue) and TE21 Homo sapiens (yellow).
Acyl carrier proteins
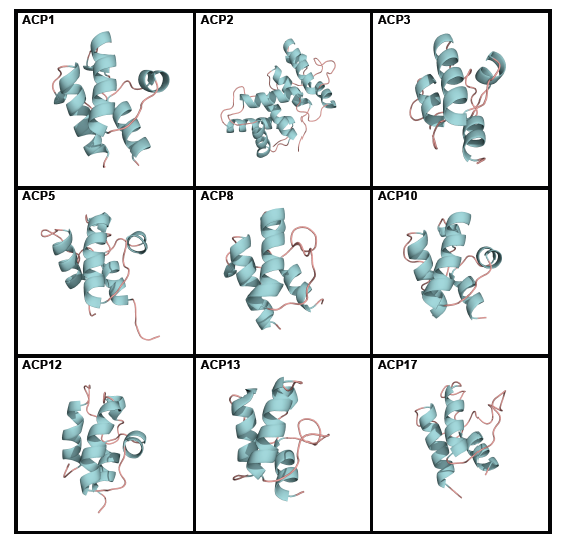
Acyl carrier proteins have very variable primary structures, and therefore they can be classified into families ACP1 to ACP17 (Figure 6), family ACP14 having been deleted [6] (Table 1). Most acyl carrier proteins have 70 to 100 amino acid residues, but members of family ACP2 have longer chains. Commonly found are three parallel α-helices, usually but not always accompanied by a crosswise α-helix (Figure 7). Members of some families are freestanding, while others are part of longer fatty acid synthase and polyketide synthase chains (Table 1). Members of individual acyl carrier protein families are more likely than members of the eight enzyme groups that are part of the fatty acid synthesis cycle to be made by members of single life kingdoms, usually bacterial but sometimes eukaryotic.
Figure 6. Tertiary structures of single members of acyl carrier protein (ACP) families. ACP1 Thermus thermophilus, ACP2 Saccharomyces cerevisiae, ACP3 Homo sapiens, ACP5 Streptomyces roseofulvus, ACP8 Aspergillus parasiticus, ACP10 Brevibacillus parabrevis, ACP12 Escherichia coli, ACP13 E. coli, and ACP17 Lactobacillus casei. Reproduced with permission from ref. 6. Copyright 2012, The Protein Society.
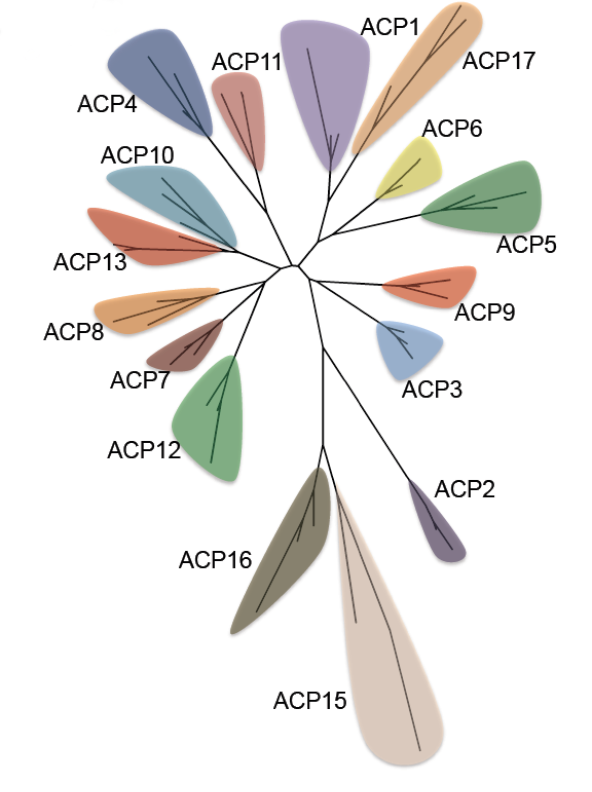
Figure 7. Phylogenetic tree of acyl carrier protein families. Reproduced with permission from ref. 6. Copyright 2012, The Protein Society.
Computation has established that ACP4 members have folds similar to those of other acyl carrier protein families whose folds have been experimentally determined [6]. However, members of families ACP15 and ACP16 appear by computation to have different tertiary structures, in agreement with their having more divergent primary structures (Figure 6) and different roles (they are associated with enzymes that decrease rather than increase the sizes of substrates and they do not use the same 4’-phosphopantetheine prosthetic group present in other acyl carrier proteins).
Fatty acid and polyketide synthesis complexes
It has been noted above and in Table 1 that the members of some fatty acid synthesis enzyme families are covalently linked parts of fatty acid and polyketide synthesis complexes. More specifically, these complexes, found in some bacteria, fungi, animals, and slime molds, allow growing fatty acid chains, activated by acyl carrier protein, to be passed sequentially from one fatty acid synthesis enzyme to the next without randomly diffusing among them.
Of the sixteen acyl carrier protein families, members of ACP1, ACP4, ACP5, ACP15, ACP16, and ACP17 are freestanding (Table 1) and engage in what is known as Type II synthesis. ACP1 proteins are produced by bacteria, fungi, animals, and plants, and they engage with many fatty acid synthesis enzymes. ACP4 and ACP5 members are produced by various bacteria and are involved with polyketide synthesis. Members of ACP15 and ACP16 are also produced by various bacteria, and they are associated with malonate decarboxylase and citrate lyase, respectively. ACP17 is composed of D-alanyl carrier proteins.
The remaining acyl carrier proteins covalently linked to enzymes take part in Type I fatty acid synthesis and other related activities (Table 1). ACP2 and ACP3 proteins help to produce fatty acids and are made by fungi and animals, respectively. Members of ACP6 through ACP11 are engaged in polyketide synthesis and are produced by mycobacteria (ACP6), fungi (ACP7, ACP8, and ACP11), and slime molds (ACP9). The very large number of ACP10 proteins are made by various bacteria as well as by fungi. ACP12 and ACP13 members are made by various bacteria and are linked to isochoromatases, peptide synthetases, phenyloxazoline synthases (ACP12), enterobactin synthases, and chromophore lyases (ACP13)
Of the eight fatty acid synthesis enzyme groups, six (acyltransferases, ketoacyl synthases, ketoacyl reductases, hydroxyacyl dehydratases, enoyl reductases, and thioesterases) have families whose members are part of multi-enzyme fatty acid and polyketide synthases (Table 1). One acyltransferase family (AT1) has domains in various synthases produced by bacteria, fungi, and animals, as well as freestanding members. Members of the very large KS3 family are both freestanding and are parts of polyketide and fatty acid synthases produced by bacteria, animals, fungi, and slime molds. Among the ketoacyl reductases, bacterial and fungal forms of KR3 are found in fatty acid synthases, while bacterial, fungal, and animal forms of KR4 are in polyketide and fatty acid synthases. Hydroxyacyl dehydratases in HD3 are part of fatty acid synthases produced by bacteria, fungi, and animals, and those in HD4 are in polyketide and fatty acid synthases made by bacteria and animals. ER3 has enoyl reductase domains in fatty acid synthases produced by bacteria and fungi, while enoyl reductase domains in ER4 are located in bacterial, fungal, animal, and slime mold polyketide synthases. Finally, families TE16, TE17, and TE18 among the 25 thioesterase families contribute domains to multi-enzyme synthases. TE16 members are part of amino acid adenylation proteins, polyketide, fatty acid, and nonribosomal peptide synthases, and enterobactin synthase component F made by bacteria, fungi, and animals. Bacterial polyketide synthases have TE17 domains. TE18 domains are found in bacterial amino acid adenylation proteins and nonribosomal peptide synthases and in animal S-acyl fatty acid synthases.
A note in passing
Archaeal membranes contain phospholipids that have hydrophobic chains bound to L-glycerol with ether links rather than the chains found in bacteria and eukaryota bound to D-glycerol by ester links. Furthermore, archaeal chains are branched with methyl groups at regular intervals, based on isoprenoid chemistry, while bacterial and eukaryotic organisms have aryl chains with straight-chain fatty acids. Despite this, many archaea have one or more of the enzymes (or acyl carrier protein) found in the fatty acid synthesis cycle. Chen used the ThYme database in 2011 to find that of the 155 archaeal species tabulated then, 33 had one cycle member, 28 had two, 22 had three, 17 had four, 26 had five, 27 had six, one had eight, and one had all nine [10].
References
- Cantu, D.C., Chen, Y., Lemons, M.L. and Reilly, P.J. ThYme: A database for thioester-active enzymes. Nucleic Acids Res., 39, D342–D346 (2011). DOI: 10.1093/nar/gkq1072. http://www.enzyme.cbirc.iastate.edu.
- Chen, Y., Elizondo-Noriega, A., Cantu, D.C. and Reilly, P.J. Structural classification of biotin carboxyl carrier proteins. Biotechnol. Lett., 34, 1869–1875 (2012). DOI: 10.1007/s10529-012-0978-4.
- Chen, Y., Kelly, E.E., Masluk, R.P., Nelson, C.L., Cantu, D.C. and Reilly, P.J. Structural classification and properties of ketoacyl synthases. Protein Sci., 20, 1659–1667 (2011). DOI: 10.1002/pro.712.
- Cantu, D.C., Dai, T., Beversdorf, Z.S. and Reilly, P.J. Structural classification and properties of ketoacyl reductases, hydroxyacyl dehydratases, and enoyl reductases. Protein Eng. Des. Sel., 25, 803–811 (2012). DOI: 10.1093/protein/gzs050.
- Cantu, D.C., Chen, Y. and Reilly, P.J. Thioesterases: A new perspective based on their primary and tertiary structures. Protein Sci., 19, 1281–1295 (2010). DOI: 10.1002/pro.417.
- Cantu, D.C., Forrester, M.J., Charov, K. and Reilly, P.J. Acyl carrier protein structural classification and normal mode analysis. Protein Sci., 21, 655–666 (2012). DOI: 10.1002/pro.2050.
- Phan, N.N., Lee, Y.K. and Reilly, P.J. Fatty acid synthesis enzyme clans. Biotechnol. Lett., 37, 417–427 (2015). DOI: 10.1007/s10529-014-1687-y.
- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). http://www.chem.qmul.ac.uk/iubmb/enzyme/.
- Lombard, J., Moreira, D. Early evolution of the biotin-dependent carboxylase family. BMC Evol. Biol., 11, 232 (2011). DOI: 10.1186/1471-2148-11-232.
- Chen, Y. Department of Chemical and Biological Engineering, Iowa State University. Unpublished research (2011).