Polyhydroxyalkanoates
The Author: Bernd HA Rehm, Institute of Fundamental Sciences and MacDiarmid Institute for Advanced Materials and Nanotechnology,, Massey University, Palmerston North, New Zealand
Introduction
PHAs (polyhydroxyalkanoates = biopolyesters) were discovered at the beginning of the 20th century by Lemoigne (1926) when observing poly(3-hydroxybutyrate) (PHB) granules inside the Gram-positive bacterium Bacillus megaterium [1].
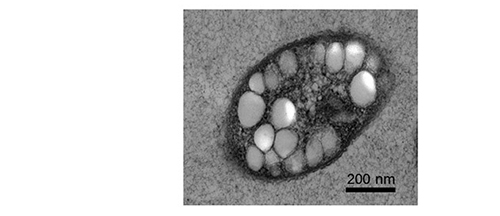
PHAs are composed of hydroxy fatty acids and represent a complex class of intracellular storage polymers synthesized by various bacteria and archaea. PHAs are produced in the presence of excess carbon source while growth is inhibited due to limited nutrient availability. PHAs are deposited as water-insoluble granules inside the cells (Figure 1). However, under carbon starvation conditions granule-associated PHA depolymerizing enzymes degrade the PHA to provide carbon and energy.
Figure 1: Electron microscopy image of bacterial cell accumulating PHA granules
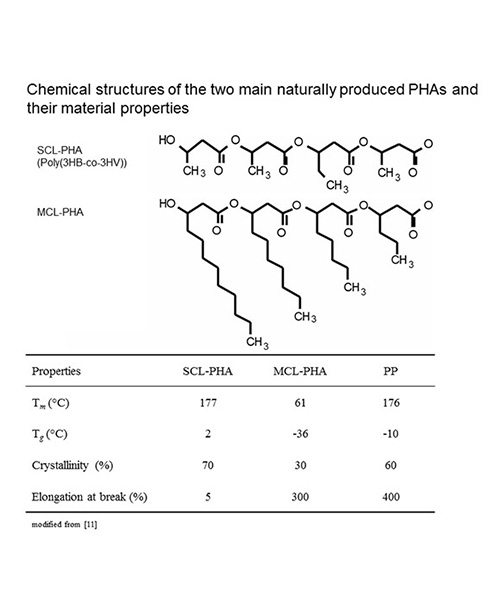
Most prokaryotic microorganisms produce PHB which is composed of (R)-3-hydroxybutyrate with a molecular weight ranging from about 500000 to several millions. Purified PHB is a crystalline, rather brittle thermoplastic material which has been considered for bulk application to replace commodity oil-based products (Table 1). The second major class of naturally produced PHAs is composed of medium-chain length (MCL) (R)-3-hydroxyfatty acids (6-14 carbon atoms) with molecular weights ranging from about 100000 – 500000. These PHAs are mainly produced by fluorescent pseudomonads. Purified MCL-PHAs show less crystallinity and hence enhanced elastomeric properties (Table 1). More than 150 constituents had been demonstrated to be incorporated into PHAs. Decades of PHA research were dedicated to understanding the biosynthesis pathways towards the production of these biopolymers as well as their material properties and potential applications [2, 3]. Overall, the thermoplastic properties, biodegradability and biocompatibility make these renewable materials suitable for several applications in packaging industry, medicine, pharmacy, agriculture and food industry or as raw materials for the synthesis of enantiomerically pure chemicals ((R)-3-hydroxyfatty acids) [4, 5].
Worldwide, several companies were formed to commercialize PHAs as biodegradable plastics [5]. PHAs have been marketed as renewable and environmentally friendly bioplastics. However, the commercial success of PHA bioplastics is still hampered by their high production costs, poor material properties and processability when compared to the well-established oil-based materials.
In addition to the high molecular weight PHAs deposited as intracellular storage granules, a different form of PHAs with lower molecular weight and complexed with other macromolecules (hence termed c-PHB) had been discovered in prokaryotic and eukaryotic cells [6, 7]. c-PHBs are only found in low concentrations intracellularly or are located in the membranes of cells. c-PHB complexed with calcium polyphosphate and embedded in bacterial membranes might form ion channels and also may be involved in competence of bacterial cells i.e. for their ability to take up DNA [8, 9]. Only limited information is available about c-PHB biosynthesis. However, in a more recent study the protein YdcS was identified in Escherichia coli as showing PHB synthase activity [10]. Primary structure analysis of YdcS suggested that it belongs to the α-/β-hydrolase superfamily that comprises PHB synthases (known for storage PHB synthesis), lipases, and esterases. YdcS was proposed as c-PHB synthase.
Polyhydroxyalkanoate biosynthesis
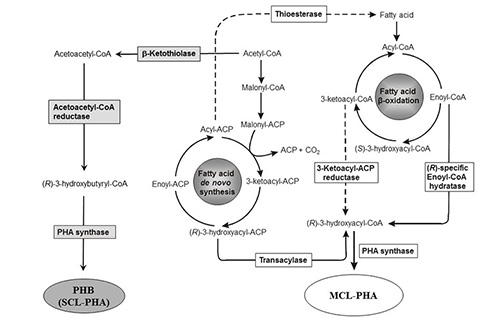
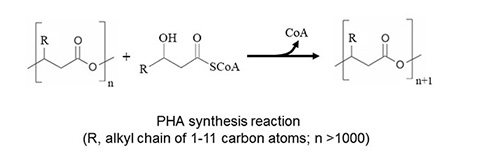
The key enzymes of PHA biosynthesis are the PHA synthases that catalyze the processive formation of the PHA chain [11]. Various metabolic routes are realized towards the synthesis of the activate PHA precursor, (R)-3-hydroxyacyl-coenzyme A (CoA), which serves as substrate for the PHA synthase [12] (Figure 2).
Figure 2: PHA biosynthesis pathways. Dashed lines show examples of engineered pathway steps toward the synthesis of tailored PHAs (modified according to [13]).
The biosynthesis of PHB begins with the condensation of two acetyl-CoA molecules catalyzed by the b–ketothiolase (PhaA) resulting in the formation of acetoacetyl-CoA which is then reduced to (R)-3-hydroxybutyryl-CoA by the (R)-specific acetoacetyl-CoA reductase (PhaB) [13]. (R)-3-Hydroxybutyryl-CoA is the activated precursor of PHB and substrate for the PHA synthase (PhaC) (Scheme 1). The genes encoding PHB biosynthesis proteins are often co-localized and organized in an operon [11] (Table 2). However, PHAs composed of MCL (R)-3-hydroxyfatty acids are synthesized diverting intermediates of fatty acid metabolism to (R)-3-hydroxyacyl-CoA and thus towards MCL-PHA (Figure 2). If the carbon source is oxidized to acetyl-CoA, excluding its formation by the fatty acid b-oxidation pathway, then intermediates of fatty acid de novo biosynthesis become the precursors for MCL-PHA biosynthesis and whose conversion is catalysed by the transacylase PhaG.
This specific transacylase is involved in the transfer of the (R)-3-hydroxyacyl moiety of the respective ACP (acyl carrier protein) thioester to CoA [14, 15] (Figure 2). PhaG has been employed for metabolic engineering of MCL-PHA biosynthesis in order to obtain tailor-made biopolyester [14, 16-21]. If the carbon source is oxidized through the fatty acid b-oxidation pathway, then the (R)-specific enoyl-CoA hydratase (PhaJ) catalyses the oxidation of enoyl-CoA to (R)-3-hydroxyacyl-CoA [22] (Figure 2). (R)-3-Hydroxyacyl-CoA is substrate for the PHA synthase (PhaC) and the direct precursor of PHA biosynthesis. PhaJ has been also employed for metabolic engineering of MCL-PHA biosynthesis in order to obtain tailor-made biopolyester [23-26]. The genes encoding for the metabolically linking enzymes (PhaG, PhaJ) are not colocalized with the PHA synthase gene, but are co-regulated (Table 2). Metabolic engineering had been extensively explored to obtain PHAs with defined composition and desired material properties suitable for medical and industrial applications [27, 28].
PHA synthases
Classes of PHA synthases
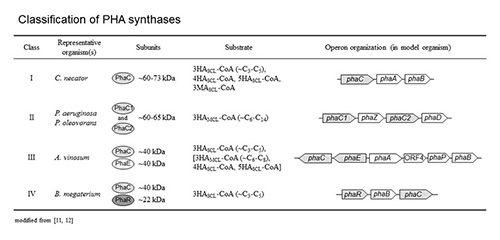
PHA synthases can be divided into four main classes with respect to substrate specificity and subunit composition (Table 2). The PHA synthase belonging to classes I or II form a homodimer as soon as substrate is made available and polymerisation starts [29, 30] (Figure 3). The synthases that are actively forming the PHA chain assemble to a small PHA granule with hydrophobic PHA chains in the core and the synthases at the surface [13, 31] (Figures 4 and 5). The PHA synthases remain covalently attached to the core and move apart while the PHA granule is increasing in size [32, 33] (Figure 5).
Figure 3: Proposed reaction mechanism of PHA synthases (modified according to [11])
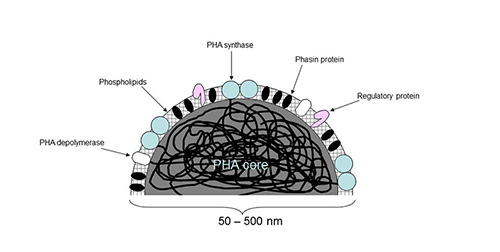
Figure 4: Schematic view of a PHA granule (modified according to [2])
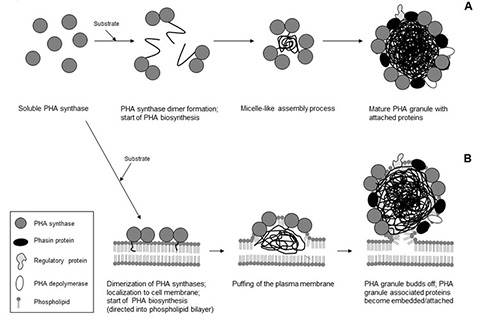
Figure 5: Models of PHA granules formation. A, Micelle model; B, Budding model (modified according to [3])
Eight conserved amino acid residues, six conserved blocks and a conserved α/β-hydrolase fold region were identified based on multiple alignment as well as a conserved domain database search of PHA synthase primary structures [11, 34]. Amino acid residues cysteine-319, aspartate-480 and histidine-508 of the model class I PHA synthase from Ralstonia eutropha are conserved in all PHA synthases and are required for covalent catalysis [11, 35, 36]. Cysteine-319 is the proposed catalytic nucleophile that is activated by the general base catalyst histidine-508 (Figure 3).
PHA synthases of class I are composed of one subunit (PhaC) of 60-73 kDa and synthesize PHA composed of short-chain-length monomers (SCL-PHA) [11]. Class II PHA synthases are found in fluorescent pseudomonads and also comprise only one type of subunit (PhaC) [37]. In contrast to class I PHA synthases, the preferred substrate is (R)-3-hydroxyacyl-CoA composed of 6 to 14 carbon atoms. Dissimilar to class I and II synthases, class III PHA synthases are composed of two subunits: one PhaC subunit of about 40 kDa which primary structure exhibits strong similarity to class I and class II PHA synthases and a PhaE subunit (about 40 kDa) with no similarity to PHA synthases [38, 39]. Class III and class IV PHA synthases synthesize SCL-PHAs. Class IV PHA synthases can be found in bacteria belonging to the genus Bacillus and are composed of a PhaC subunit (about 40 kDa) and a smaller PhaR subunit of about 20 kDa [40]. Investigations of the substrate specificity of PHA synthases have been mostly carried out in vitro or in vivo in recombinant E. coli. Interestingly, in vitro studies showed a narrow substrate specificity [41, 42] whereas the substrate specificity in E. coli was rather broad [43-45].
PHA synthases: structure-function
Currently no PHA synthase structural information based on X-ray crystallography or NMR spectroscopy is available. PHA synthase amino acid sequence alignments revealed six conserved regions as well as eight conserved amino acids [11]. While N-terminal regions of most PHA synthases are less conserved, the C-terminus shows a higher level of conservation. Deletion mutagenesis showed that the first 100 amino acid residues of the PHA synthase of R. eutropha were dispensable, i.e. not required for enzyme activity [34, 46]. However, single amino acid substitution at position four of the N-terminus of the PHA synthase from R. eutropha led to increased copy numbers of PHA synthase coinciding with enhanced PHB accumulation [47]. These data suggested some functional role of the variable N-terminus of PHA synthases. The more conserved C-terminal region (~40 C-terminal amino acid residues) of the class I and class II PHA synthases is composed of mainly hydrophobic amino acid residues. Since deletion of only five amino acid residues of the C-terminus of class I PHA synthase from R. eutropha did already result in loss of enzyme activity, this region seems to play an important role in enzyme functions, presumably by anchoring the PHA synthase to the hydrophobic core of the PHA granule [34]. In class III and IV PHA synthases, the subunits PhaE or PhaR exhibit a very hydrophobic C-terminal region and hence might be involved in anchoring the heterodimeric PHA synthase to the PHA core of the granules.
PHA synthases contain a conserved lipase box (GX[S/C]XG) in which serine at the active site is substituted by a cysteine. Moreover, conserved-domain-homology searches suggested that PHA synthases belong to the family of α/β-hydrolases containing conserved structural features as found for example in lipases. Threading models of the class I (R. eutropha) [34], of class II (P. aeruginosa) [48], and of class III (A. vinosum) [49] PHA synthases showed the α/β-hydrolase fold with its conserved catalytic triad motif (nucleophile-acid-base). A further resemblance to lipases is the increased activity of granule-bound PHA synthase compared to the soluble counterpart [50] which suggested interfacial activation that is characteristic for lipases but at the lipid-water interface.
Reaction mechanism
The catalytic triad residues, cysteine, aspartate and histidine (C149, D302 and H331 in A. vinosum or C319, D480 and H508 in R. eutropha), of the PHA synthase were proposed to be directly involved in the reaction mechanism. Structural models also suggested that these residues constitute the active site [11, 48, 51, 52]. It was also shown for class I and II synthase that substrate binding induces dimerization suggesting the involvement of two activated thiol groups each contributed by one subunits [29, 30]. Activation of active site cysteines by histidines mediates the nucleophilic attack on the thioester bond of two (R)-3-hydroxyacyl-CoA substrates. After this initiation step, both thiol groups form a thioester bond with 3-hydroxy-fatty acid (Figure 3). The elongation step involves the conserved aspartate of one subunit which activates the hydroxyl group of the bound 3-hydroxy-fatty acid that in turn attacks the thioester bond between the cysteine and the hydroxyl-fatty acid of the second subunit. Hence the nascent PHA chain remains bound to one subunit of the active dimer and the free thiol group of the other subunit can bind substrate i.e. the next building block of the PHA chain. Similar to the mechanism of the fatty acid synthase, the growing PHA chain moves from one active site to the other while being extended by one building block [53]. Alternatively as found in α/β-hydrolases only one active site might be involved in covalently binding substrate while the hydroxyacyl-CoA monomers binds non-covalently to synthase.
In vivo assembly of PHA granules
PHA granules are water-insoluble spherical inclusions inside the bacterial cell, and their detailed structure and composition has been subject to extensive investigations commencing decades ago. Early studies on PHB granule composition showed that protein and lipid were present in addition to the major component of PHB [54]. Twenty years later, high resolution 13C-NMR spectroscopy analysis of live cells showed that PHB is predominantly in a mobile amorphous state in the granule core as opposed to the solid (i.e. crystallised) state after extraction [55]. The same authors also suggested that water molecules are present within the PHB core and serve as plasticizer for the polymer. The PHA core is surrounded by a boundary layer that is composed of a phospholipid-monolayer [56] plus proteins [57] (Figure 4). These proteins that are attached to the PHA core can be subdivided into structural proteins, the phasins, the PHA synthase, the intracellular PHA depolymerase, and regulator proteins [58, 59]. Contrast-variation small-angle neutron scattering (SANS) has confirmed the presence of a phospholipid-monolayer [60]. Atomic force microscopy (AFM) studies provided further evidence for a phospholipid monolayer boundary and additionally suggested complex structures such as cytoskeleton-like phasin based scaffold at the granule surface [61, 62].
Two models of PHA granule assembly are currently being discussed: the “micelle” and the “budding” model (Figure 5). In the “micelle” model, the randomly distributed cytosolic PHA synthase begins to synthesize the PHA chain converting the soluble synthase into an amphipathic PHA:enzyme complex which self-organizes into a small micelle with a PHA core and the PHA synthase at the surface. Continuous PHA synthesis causes the PHA inclusion to increase in size and phospholipids, as well as proteins, become incorporated into the PHA granule surface. The strongest support for the micelle model is provided by in vitro PHA granule formation and by in vivo studies using fluorescently labelled PHA synthase [50, 63].
In contrast, the “budding” model requires the PHA synthase to locate to the cytoplasmic membrane thereby catalysing synthesis of PHA into the hydrophobic space between the phospholipid layers which ultimately results in the PHA granules budding off from the membrane. Budding would coat the granules with a phospholipid monolayer in which proteins become embedded.
Time-course studies on PHB granule formation in R. eutropha using transmission electron microscopy (TEM) showed that PHA granule formation seems to occur in the centre of the cell at so-called mediation elements which might serve as nucleation sites for PHA granule assembly [64].
Peters and Rehm (2005) used fluorescence microscopy with a fluorescently-labelled PHA synthase that showed that emerging PHA granules located to the cell poles and to the mid-cell for the creation of (?) future cell poles [63]. Neither the Z-ring (septum) formation nor PHA chain synthesis impacted localization of the PHA synthase within the cell but chromosome condensation had an influence [63, 65].
Jendrossek [66] applied fluorescence microscopy using Nile Red staining of PHA granules and fluorescently-labelled phasins which showed that emerging PHA granules were often found close to the cell poles as well as to the cytoplasmic membrane.
Further PHA-accumulating bacteria were studied which revealed a common localisation either close to the cytoplasmic membrane or the cell poles more in alignment with budding model. However, since granules are formed in vitro when purified, soluble PHA synthase is exposed to substrate, a membrane environment does not seem to be essential for granule assembly which is in favour of the micelle model.
PHA granules as functionalized biobeads
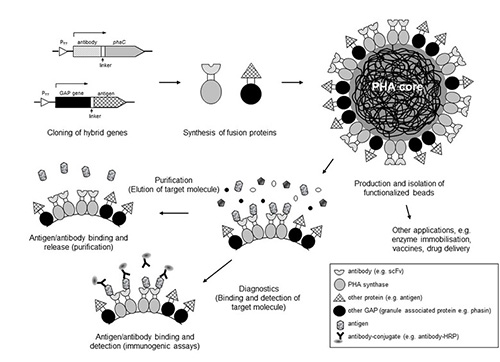
After disruption of cells that are accumulating, PHA granules could be isolated, purified and stably maintained as spherical shell-core particles outside the cell. This property has led to extensive protein engineering approaches targeting proteins specifically interacting with the PHA core such as e.g. PHA synthases or phasins. The proteins have been engineered by translationally fusing or inserting protein-based functions such as e.g. binding domains, antigens, enzymes or fluorescent proteins [31, 59, 67] (Figure 6). These functions are localized to the surface of isolated PHA granules and their respective uses, such as in bioseparations, vaccines, enzyme immobilisation or diagnostics, was demonstrated [68-71]. The engineering of surface functions into PHA granules has opened up a new avenue for the potential commercialisation of PHAs.
Figure 6: Funtionalization of PHA granules as biobeads (modified according to [59]).
References
- Lemoigne, M., Produits de deshydration et de polymerisation de l’acide β-oxybutyric. Bull. Soc. Chim. Biol., 1926. 8: p. 770-782.
- Rehm, B., Bacterial polymers: biosynthesis, modifications and applications. Nature Reviews Microbiology, 2010. 8(8): p. 578-592.
- Rehm, B.H., Biogenesis of microbial polyhydroxyalkanoate granules: a platform technology for the production of tailor-made bioparticles. Curr Issues Mol Biol, 2007. 9(1): p. 41-62.
- Lee, S.Y. and Y. Lee, Metabolic engineering of Escherichia coli for production of enantiomerically pure (R)-(--)-hydroxycarboxylic acids. Appl Environ Microbiol, 2003. 69(6): p. 3421-6.
- Urtuvia, V., et al., Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int J Biol Macromol, 2014. 70: p. 208-13.
- Reusch, R.N., E.M. Bryant, and D.N. Henry, Increased poly-(R)-3-hydroxybutyrate concentrations in streptozotocin (STZ) diabetic rats. Acta Diabetol, 2003. 40(2): p. 91-4.
- Reusch, R.N. and H.L. Sadoff, Putative structure and functions of a poly-beta-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc Natl Acad Sci U S A, 1988. 85(12): p. 4176-80.
- Addison, C.J., S.H. Chu, and R.N. Reusch, Polyhydroxybutyrate-enhanced transformation of log-phase Escherichia coli. Biotechniques, 2004. 37(3): p. 376-8, 380, 382.
- Castuma, C.E., et al., Inorganic polyphosphates in the acquisition of competence in Escherichia coli. J Biol Chem, 1995. 270(22): p. 12980-3.
- Dai, D. and R.N. Reusch, Poly-3-hydroxybutyrate synthase from the periplasm of Escherichia coli. Biochem Biophys Res Commun, 2008. 374(3): p. 485-9.
- Rehm, B., Polyester synthases: natural catalysts for plastics. Biochemical Journal, 2003. 376: p. 15-33.
- Rehm, B. and A. Steinbuchel, Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. International Journal of Biological Macromolecules, 1999. 25(1-3): p. 3-19.
- Rehm, B.H., Genetics and biochemistry of polyhydroxyalkanoate granule self-assembly: The key role of polyester synthases. Biotechnol Lett, 2006. 28(4): p. 207-13.
- Hoffmann, N., et al., Biochemical characterization of the Pseudomonas putida 3-hydroxyacyl ACP:CoA transacylase, which diverts intermediates of fatty acid de novo biosynthesis. J Biol Chem, 2002. 277(45): p. 42926-36.
- Rehm, B.H., N. Kruger, and A. Steinbuchel, A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The PHAG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme a transferase. J Biol Chem, 1998. 273(37): p. 24044-51.
- Zheng, Z., et al., Production of 3-hydroxydecanoic acid by recombinant Escherichia coli HB101 harboring phaG gene. Antonie Van Leeuwenhoek, 2004. 85(2): p. 93-101.
- Matsumoto, K., et al., Cloning and characterization of the Pseudomonas sp. 61-3 phaG gene involved in polyhydroxyalkanoate biosynthesis. Biomacromolecules, 2001. 2(1): p. 142-7.
- Matsumoto, K., et al., Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) copolymer from sugars by recombinant Ralstonia eutropha harboring the phaC1Ps and the phaGPs genes of Pseudomonas sp. 61-3. Biomacromolecules, 2001. 2(3): p. 934-9.
- Hoffmann, N., A. Steinbuchel, and B.H. Rehm, Homologous functional expression of cryptic phaG from Pseudomonas oleovorans establishes the transacylase-mediated polyhydroxyalkanoate biosynthetic pathway. Appl Microbiol Biotechnol, 2000. 54(5): p. 665-70.
- Hoffmann, N., A. Steinbuchel, and B.H. Rehm, The Pseudomonas aeruginosa phaG gene product is involved in the synthesis of polyhydroxyalkanoic acid consisting of medium-chain-length constituents from non-related carbon sources. FEMS Microbiol Lett, 2000. 184(2): p. 253-9.
- Fiedler, S., A. Steinbuchel, and B.H. Rehm, PhaG-mediated synthesis of Poly(3-hydroxyalkanoates) consisting of medium-chain-length constituents from nonrelated carbon sources in recombinant Pseudomonas fragi. Appl Environ Microbiol, 2000. 66(5): p. 2117-24.
- Fukui, T., N. Shiomi, and Y. Doi, Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol, 1998. 180(3): p. 667-73.
- Tsuge, T., et al., Alteration of chain length substrate specificity of Aeromonas caviae R-enantiomer-specific enoyl-coenzyme A hydratase through site-directed mutagenesis. Appl Environ Microbiol, 2003. 69(8): p. 4830-6.
- Lu, X., et al., Enhanced production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) via manipulating the fatty acid beta-oxidation pathway in E. coli. FEMS Microbiol Lett, 2003. 221(1): p. 97-101.
- Fiedler, S., A. Steinbuchel, and B.H. Rehm, The role of the fatty acid beta-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans and of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch Microbiol, 2002. 178(2): p. 149-60.
- Fukui, T., et al., Co-expression of polyhydroxyalkanoate synthase and (R)-enoyl-CoA hydratase genes of Aeromonas caviae establishes copolyester biosynthesis pathway in Escherichia coli. FEMS Microbiol Lett, 1999. 170(1): p. 69-75.
- Madison, L.L. and G.W. Huisman, Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev, 1999. 63(1): p. 21-53.
- Aldor, I.S. and J.D. Keasling, Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol, 2003. 14(5): p. 475-83.
- Rehm, B.H., et al., Matrix-assisted in vitro refolding of Pseudomonas aeruginosa class II polyhydroxyalkanoate synthase from inclusion bodies produced in recombinant Escherichia coli. Biochem J, 2001. 358(Pt 1): p. 263-8.
- Wodzinska, J., et al., Polyhydroxybutyrate synthase: Evidence for covalent catalysis. JOURNAL OF THE AMERICAN CHEMICAL SOCIETY, 1996. 118(26): p. 6319-6320.
- Draper, J.L. and B.H. Rehm, Engineering bacteria to manufacture functionalized polyester beads. Bioengineered, 2012. 3(4): p. 203-8.
- Hezayen, F.F., A. Steinbuchel, and B.H. Rehm, Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56. Arch Biochem Biophys, 2002. 403(2): p. 284-91.
- Peters, V. and B.H.A. Rehm, In vivo enzyme immobilisation using engineered PHA synthase. Appl Environ Microbiol, 2006. in press.
- Rehm, B.H., et al., Molecular characterization of the poly(3-hydroxybutyrate) (PHB) synthase from Ralstonia eutropha: in vitro evolution, site-specific mutagenesis and development of a PHB synthase protein model. Biochim Biophys Acta, 2002. 1594(1): p. 178-90.
- Jia, Y., et al., Mechanistic studies on class I polyhydroxybutyrate (PHB) synthase from Ralstonia eutropha: class I and III synthases share a similar catalytic mechanism. Biochemistry, 2001. 40(4): p. 1011-9.
- Gerngross, T.U., et al., Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry, 1994. 33(31): p. 9311-20.
- Timm, A. and A. Steinbuchel, Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol, 1990. 56(11): p. 3360-7.
- Liebergesell, M. and A. Steinbuchel, Cloning and molecular analysis of the poly(3-hydroxybutyric acid) biosynthetic genes of Thiocystis violacea. Appl Microbiol Biotechnol, 1993. 38(4): p. 493-501.
- Liebergesell, M. and A. Steinbuchel, Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur J Biochem, 1992. 209(1): p. 135-50.
- McCool, G.J. and M.C. Cannon, Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol, 1999. 181(2): p. 585-92.
- Anderson, A.J., G.W. Haywood, and E.A. Dawes, Biosynthesis and composition of bacterial poly(hydroxyalkanoates). Int J Biol Macromol, 1990. 12(2): p. 102-5.
- Yuan, W., et al., Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch Biochem Biophys, 2001. 394(1): p. 87-98.
- Langenbach, S., B.H. Rehm, and A. Steinbuchel, Functional expression of the PHA synthase gene phaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett, 1997. 150(2): p. 303-9.
- Qi, Q., B.H. Rehm, and A. Steinbuchel, Synthesis of poly(3-hydroxyalkanoates) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett, 1997. 157(1): p. 155-62.
- Antonio, R.V., A. Steinbuchel, and B.H. Rehm, Analysis of in vivo substrate specificity of the PHA synthase from Ralstonia eutropha: formation of novel copolyesters in recombinant Escherichia coli. FEMS Microbiol Lett, 2000. 182(1): p. 111-7.
- Schubert, P., N. Kruger, and A. Steinbuchel, Molecular analysis of the Alcaligenes eutrophus poly(3-hydroxybutyrate) biosynthetic operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and identification of the promoter. J Bacteriol, 1991. 173(1): p. 168-75.
- Normi, Y.M., et al., Characterization and properties of G4X mutants of Ralstonia eutropha PHA synthase for poly(3-hydroxybutyrate) biosynthesis in Escherichia coli. Macromol Biosci, 2005. 5(3): p. 197-206.
- Amara, A.A. and B.H. Rehm, Replacement of the catalytic nucleophile cysteine-296 by serine in class II polyhydroxyalkanoate synthase from Pseudomonas aeruginosa-mediated synthesis of a new polyester: identification of catalytic residues. Biochem J, 2003. 374(Pt 2): p. 413-21.
- Jia, Y., et al., Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry, 2000. 39(14): p. 3927-36.
- Gerngross, T.U. and D.P. Martin, Enzyme-catalyzed synthesis of poly[(R)-(-)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc Natl Acad Sci U S A, 1995. 92(14): p. 6279-83.
- Li, P., S. Chakraborty, and J. Stubbe, Detection of covalent and noncovalent intermediates in the polymerization reaction catalyzed by a C149S class III polyhydroxybutyrate synthase. Biochemistry, 2009. 48(39): p. 9202-11.
- Tian, J., A.J. Sinskey, and J. Stubbe, Detection of intermediates from the polymerization reaction catalyzed by a D302A mutant of class III polyhydroxyalkanoate (PHA) synthase. Biochemistry, 2005. 44(5): p. 1495-503.
- Witkowski, A., A.K. Joshi, and S. Smith, Characterization of the interthiol acyltransferase reaction catalyzed by the beta-ketoacyl synthase domain of the animal fatty acid synthase. Biochemistry, 1997. 36(51): p. 16338-44.
- Griebel, R., Z. Smith, and J.M. Merrick, Metabolism of poly-beta-hydroxybutyrate. I. Purification, composition, and properties of native poly-beta-hydroxybutyrate granules from Bacillus megaterium. Biochemistry, 1968. 7(10): p. 3676-81.
- Barnard, G.N. and J.K. Sanders, The poly-beta-hydroxybutyrate granule in vivo. A new insight based on NMR spectroscopy of whole cells. J Biol Chem, 1989. 264(6): p. 3286-91.
- Boatman, E.S., Observations on the Fine Structure of Spheroplasts of Rhodospirillum Rubrum. J Cell Biol, 1964. 20: p. 297-311.
- Pieper-Furst, U., et al., Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J Bacteriol, 1995. 177(9): p. 2513-23.
- Jendrossek, D. and D. Pfeiffer, New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol, 2014. 16(8): p. 2357-73.
- Grage, K., et al., Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules, 2009. 10(4): p. 660-9.
- Russell, R.A., et al., Production and use of deuterated polyhydroxyoctanoate in structural studies of PHO inclusions. J Biotechnol, 2007. 132(3): p. 303-5.
- Dennis, D., et al., PhaP is involved in the formation of a network on the surface of polyhydroxyalkanoate inclusions in Cupriavidus necator H16. J Bacteriol, 2008. 190(2): p. 555-63.
- Dennis, D., et al., Preliminary analysis of polyhydroxyalkanoate inclusions using atomic force microscopy. FEMS Microbiol Lett, 2003. 226(1): p. 113-9.
- Peters, V. and B. Rehm, In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. Fems Microbiology Letters, 2005. 248(1): p. 93-100.
- Tian, J., A.J. Sinskey, and J. Stubbe, Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J Bacteriol, 2005. 187(11): p. 3814-24.
- Peters, V., D. Becher, and B.H. Rehm, The inherent property of polyhydroxyalkanoate synthase to form spherical PHA granules at the cell poles: the core region is required for polar localization. J Biotechnol, 2007. 132(3): p. 238-45.
- Jendrossek, D., Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol, 2009. 191(10): p. 3195-202.
- Hooks, D.O., et al., Polyhydroyxalkanoate synthase fusions as a strategy for oriented enzyme immobilisation. Molecules, 2014. 19(6): p. 8629-43.
- Hay, I.D., et al., Bioengineering of bacteria to assemble custom-made polyester affinity resins. Appl Environ Microbiol, 2015. 81(1): p. 282-91.
- Jahns, A.C. and B.H. Rehm, Immobilization of active lipase B from Candida antarctica on the surface of polyhydroxyalkanoate inclusions. Biotechnol Lett, 2015. 37(4): p. 831-5.
- Chen, S., et al., New skin test for detection of bovine tuberculosis on the basis of antigen-displaying polyester inclusions produced by recombinant Escherichia coli. Appl Environ Microbiol, 2014. 80(8): p. 2526-35.
- Parlane, N.A., et al., Production of a particulate hepatitis C vaccine candidate by an engineered Lactococcus lactis strain. Appl Environ Microbiol, 2011. 77(24): p. 8516-22.