Lipidomic Analysis of Glycerolipids
The Author: Robert C. Murphy, Department of Pharmacology, MS 8303 University of Colorado Denver, Aurora, CO 80045, U.S.A.
The classification of lipids within the glycerolipid category includes those molecules which have the 3-carbon carbohydrate glycerol as a common structural modality. An arbitrary decision was made by the LIPID MAPS consortium not to include glycerophospholipids in this category solely because of the sheer size of both the glycerolipid and the glycerophospholipid groupings [1]. Even with this LIPID MAPS definition there still remain a large number of highly important lipids found in mammalian, plant, bacterial, and archaebacterial cells (Fig. 1). See also our web pages 'A Lipid Primer'.
| Figure 1. Glycerolipid subclassification by LIPID MAPS [1] | |
| Monoradylglycerols | |
| Monoacylglycerols (MAG) Monoalkylglycerols Mono-(1Z-alkenyl)-glycerols |
|
| Diradylglycerols | |
| Diacylglycerols (DAG) 1-Alkyl,2-acylglycerols 1-Acyl,2-alkylglycerols Dialkylglycerols 1Z-Alkenylacylglycerols Di-glycerol tetraethers Di-glycerol tetraether Glycans |
|
| Triradylglycerols | |
| Triacylglycerols (TAG) Alkyldiacylglycerols Dialkylmonoacylglycerols 1Z-Alkenyldiacylglycerols Estolides |
|
| Glycosylmonoradylglycerols | |
| Glycosylmonoacylglycerols Glycosylmonoalkylglycerols |
|
| Glycosyldiradylglycerols | |
| Glycosyldiacylglycerols Glycosylalkylacylglycerols Glycosyldialkylglycerols |
|
For most cells, molecules in this group are the source of metabolic fuels that drive all the essential biochemical reactions required for normal operation of cellular events. This classification also includes molecules which signal receptor activation on the cell surface that triggers subsequent signaling cascades through various protein kinases.
Mass spectrometry played a central role in the structural characterization of all classes of glycerolipids, dating back to the studies carried out in the early days of organic mass spectrometry [2,3]. With the development of electrospray ionization (ESI) and matrix assisted laser desorption ionization (MALDI), all of the glycerolipids now can be readily analyzed as intact molecules in spite of the fact that many of them are not volatile. The remaining challenge in the analysis of glycerolipids from any biological origin is the fact that often they are present as complex mixtures of closely related molecules which differ in alkyl or fatty acyl substituents to the glycerol backbone. Therefore, means by which one can define components of these mixtures have been devised. These newer mass spectrometric ionization techniques generate intact species such as [M+NH4]+ or [M+H]+ by attachment of an ammonium cation or proton [4,5], or alkali metal ions, resulting in [M+Me]+ [6,7]. Very few studies have been reported on the formation of negative ions from these neutral lipids, but if one uses an appropriately basic reagent gas to remove a proton by atmospheric pressure chemical ionization, negative anions [M-H]- can be observed [8].
Several strategies have been developed to address the challenge of complex mixture analysis of closely related species of glycerolipids. These include reversed-phase chromatographic separation of molecular species used for the identification of triacylglycerols present in seed oils [9]. When dealing with triacylglycerols present in cell and tissue extracts, several strategies have been developed using mass spectrometry as the major means of detection of the molecules with electrospray ionization. These include prior separation of glycerolipids using normal-phase separation approaches followed by infusion of each glycerolipid subclass into a tandem mass spectrometer. For example, neutral loss scanning was used to find those molecular species of triacylglycerols which contain a specific fatty acid [10]. Shotgun lipidomic approaches have also been described for glycerolipid analysis without prior chromatographic separation (see the webpage by Han on this topic) [11].
Online normal-phase LC/MS and LC/MS/MS techniques are powerful strategies to identify glycerolipids and glycerolipid subclasses by the mass spectrometric detection of glycerolipid positive ions such as [M+NH4]+ ions [12]. The analysis of glycosylmonoradyl- and glycosyldiradylglycerols containing a highly acidic group such as sulfoquinovosyl lipids of plant origin has employed negative ion mass spectrometry in combination with normal-phase chromatographic separation to analyze the plant glycerolipids [13,14].
Profiling Molecular Species of Glycerol Lipids Using LC/MS Analysis
The separation of neutral lipids is well suited to normal-phase chromatographic separations, and thin-layer chromatography has been used extensively for this purpose. It is easy, however, to direct the effluent from high-performance liquid chromatography (HPLC) directly into a mass spectrometer and generate positive ions using electrospray ionization by adding a solvent system containing an electrolyte such as ammonium acetate to the mobile phase after HPLC separation but before the electrospray interface [12]. The addition of an electrolyte with NH4+, Na+, or Li+ in mM concentrations results in the formation of abundant positive ions of glycerolipids as [M+H]+, [M+NH4]+, [M+Na]+, or [M+Li]+, respectively.
One of the important aspects of the analysis of any biologically derived lipid class is the means by which one isolates the lipid from the biological matrix. While many approaches have employed very powerful lipid extraction protocols with chloroform/methanol containing solvent systems, this leads to isolation of not only glycerolipids but also glycerophospholipids, sphingolipids, and sterol lipids. We have employed less powerful extraction solvents with mixtures of isooctane/ethyl acetate (3:1) to efficiently extract glycerolipids but poorly extract more polar lipids such as phospholipids and certain sphingolipids. This prolongs the life of the HPLC column by having less material to elute after target glycerolipids emerge from the column.
An example of the normal-phase separation of a cellular extract of neutral lipids is shown in Figure 2. The figure shows elution of several neutral lipids including cholesterol esters (members of the sterol lipid category) as well as triacylglycerols and diacylglycerols; monoacylglycerols are also indicated during this approximately 30 min separation. The mobile phase employed for the separation (200 μL/min) was a gradient of methyl tert-butyl ether (MTBE) in isooctane starting from 5% MTBE, which was isocratic for 10 min, then from 10-25 min MTBE/isooctane (1:1; v/v) was ramped to 30% then ramped in 4 min to 90% MTBE/isooctane where it remained for 6 min before recycling. Post-column modification of the HPLC effluent was made by adding 30 μL/min, 10 mM ammonium acetate in 95:5 (v/v), acetonitrile/water via a mixing tee attached to the electrospray ion source inlet.
Figure 2. Separation of neutral lipids extracted from bone marrow-derived macrophages using normal-phase HPLC. Glycerolipids were detected using electrospray ionization and mass spectrometry [12]. The inset is an extracted ion chromatogram for a specific ion (m/z 902.8) corresponding to [M+NH4]+ adduct ions from 54:3 triacylglycerol species.
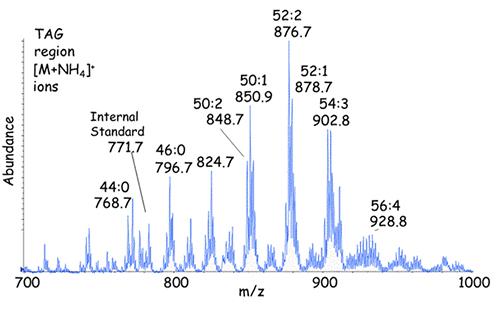
The [M+NH4]+ ions from the triacylglycerols eluting between 17.5 and 19.5 min is shown in Figure 3 with identification of the molecular species indicated using the abbreviations ACN:db (total acyl carbons:total number of fatty acyl double bonds). Any one of these individual molecular species elutes at a specific retention time in this system whereas the entire mixture of triacylglycerols appears to be a complex chromatographic peak. Elution of a specific molecular species can be seen for m/z 902.8, corresponding to 54:3-triacylglycerol as the ammonium ion adduct (Fig. 2, inset).
Figure 3. Adduct ions [M+NH4]+ from the triacylglycerols eluting from the NP-HPLC column between 17.5 and 19.5 min in Figure 2.
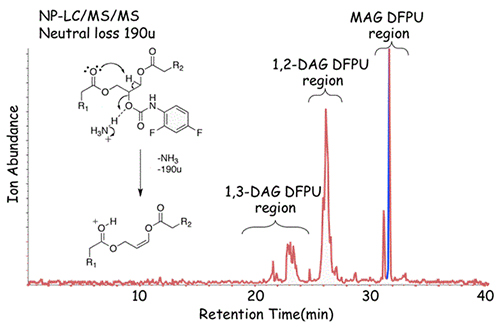
A different strategy has been developed to analyze diacylglycerols because of the unfavorable behavior of this lipid class on normal-phase silica-based columns. The acidic sites of silica catalyze fatty acyl group migration. In order to prevent this as well as facilitate chromatographic behavior, we have found that the 2,4-dinitrophenylurethane derivatives can be readily made and chromatographed [15]. This derivatization has two benefits. First of all, the stereochemistry of the diacylglycerol is fixed by blocking either the sn-1 or sn-2 position and permitting separation of 1,3- from 1,2 –diacylglycerols (Fig. 4). Perhaps of more importance is the fact that very favorable tandem mass spectrometric behavior of the 2,4-difluorophenylurethane derivatives can be used based on the common loss of 190 daltons following collisional activation of all derivatives (a possible mechanism for the formation of this ion species is indicated in Fig. 4). This permits the unique detection of diacylglycerols and monoacylglycerols as components which yield this characteristic mass loss. Therefore, neutral loss scanning can be employed to identify the diacylglycerols present in a biological extract as seen in Figure 5.
Figure 4. Separation of neutral lipids extracted from bone marrow-derived macrophages followed by derivatization with difluorophenyl isocyanate using normal-phase HPLC. The DPFU derivatives (diacylglycerols and monoacylglycerols) were detected using neutral loss scanning of the HPLC effluent with a mass offset of 190 u. The gas phase chemistry occurring with the DPFU derivative of a 1,2-diacylglycerol is illustrated in the possible reaction mechanism showing the loss of DPFU and ammonia that corresponds to 190 u [15].
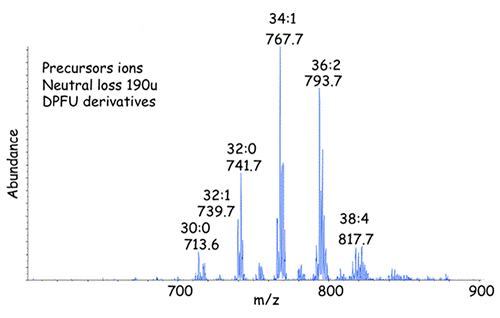
Figure 5. Precursor ions [M+NH4]+ from the 1,2-diacylglycerol-DPFU derivatives eluting from the NP-HPLC column between 24.5 and 26 min in Figure 4.
The strategies of normal-phase LC/MS and LC/MS/MS can be employed for the quantitative analysis of glycerolipids when appropriate internal standards are added to samples, as well as calibration curves for different molecular species relative to the internal standard are determined experimentally. A limitation of this technique is the accuracy of this method because of the lack of reference standard material to establish calibration curves for all molecular species of triacylglycerols and diacylglycerols present in cellular extracts. While a few of the major molecular species can be obtained pure (Avanti Polar Lipids, Alabaster, AL), these correspond to less than a few percent of the total number of triacylglycerols and diacylglycerols that are present in biological extracts.
References
- Fahy, E., Subramaniam, S., Brown, H.A., Glass, C.K., Merrill, A.H. Jr, Murphy, R.C., Raetz, C.R., Russell, D.W., Seyama, Y., Shaw, W., Shimizu, T., Spener, F., van Meer, G., VanNieuwenhze, M.S., White, S.H., Witztum, J.L. and Dennis, E.A. A comprehensive classification system for lipids. J. Lipid Res., 46, 839-861 (2005) (DOI: 10.1194/jlr.E400004-JLR200).
- Ryhage, R. and Stenhagen, E. Mass spectrometry in lipid research. J. Lipid Res., 1, 361-390 (1960).
- Lauer, W.M., Aasen, A.J., Graff, G. and Holman, R.T. Mass spectrometry of triglycerides. 1. Structural effects. Lipids, 5, 861-868 (1970).
- Duffin, K.L., Henion, J.D. and Shieh, J.J. Electrospray and tandem mass spectrometric characterization of acylglycerol mixtures that are dissolved in nonpolar solvents. Anal. Chem., 63, 1781-1788 (1991).
- McAnoy, A.M., Wu, C.C. and Murphy, R.C. Direct qualitative analysis of triacylglycerols by electrospray mass spectrometry using a linear ion trap. J. Am. Soc. Mass Spectrom., 16,1498-1509 (2005) (DOI: 10.1016/j.jasms.2005.04.017).
- Hsu, F.F. and Turk, J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple quadrupole instrument. J. Am. Soc. Mass Spectrom., 10, 587-599 (1999).
- Callender, H.L., Forrester, J.S., Ivanova, P., Preininger, A., Milne, S. and Brown, H.A. Quantification of diacylglycerol species from cellular extracts by electrospray ionization mass spectrometry using a linear regression algorithm. Anal. Chem., 79, 263-272 (2007).
- Leskinen, H.M., Suomela, J.-P. and Kallio, H.P. Quantification of triacylglycerol regioisomers by ultra-high-performance liquid chromatography and ammonia negative ion atmospheric pressure chemical ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom., 24, 1-5 (2010) (DOI: 10.1002/rcm.4346).
- Lisa, M., Holčapek, M. and Sovová, H. Comparison of various types of stationary phases in non-aqueous reversed-phase high-performance liquid chromatography–mass spectrometry of glycerol lipids in black currant oil and its enzymatic hydrolysis mixture. J. Chromatogr. A, 1216, 8371–8378 (2009) (DOI: 10.1016/j.chroma.2009.09.060).
- Murphy, R.C., James, P.F., McAnoy, A.M., Krank, J., Duchoslav, E. and Barkley, R.M. Detection of the abundance of diacylglycerol and triacylglycerol molecular species in cells using neutral loss mass spectrometry. Anal. Biochem., 366, 59-70 (2007) (DOI: 10.1016/j.ab.2007.03.012).
- Gross, R.W. and Han, X. Lipidomics in diabetes and the metabolic syndrome. Methods Enzymol., 433, 73-90 (2007).
- Hutchins, P.M., Barkley, R.M. and Murphy, R.C. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res., 49, 804-813 (2008) (DOI: 10.1194/jlr.M700521-JLR200).
- Herrero, M., Vicente, M.J., Cifuentes, A. and Ibáñez, E. Characterization by high-performance liquid chromatography/electrospray ionization quadrupole time-of-flight mass spectrometry of the lipid fraction of Spirulina platensis pressurized ethanol extract. Rapid Commun. Mass Spectrom., 21, 1729-1738 (2007) (DOI: 10.1002/rcm.3017).
- Vieler, A., Wilhelm, C., Goss, R., Süss, R. and Schiller, J. The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids, 150, 143-155 (2007) (DOI: 10.1016/j.chemphyslip.2007.06.224).
- Leiker, T.J., Barkley, R.M. and Murphy, R.C. Analysis of diacylglycerol molecular species in cellular lipid extracts by normal-phase LC-electrospray mass spectrometry. Intl. J. Mass Spectrom., 305, 103-109 (2011).