Transport and function of lipids in the plant phloem
The Author: Susanne Hoffmann-Benning, Assistant Professor at Michigan State University, East Lansing, Michigan, USA
Introduction
Lipids are essential components of plants. They provide the energy for metabolic processes, are structural components for membranes, and are important intracellular signals. In recent years a variety of lipophilic molecules have been discussed as long-distance signals as well. While some of these lipids are related to biotic stress/pathogen response and systemic acquired resistance, others are good candidates for signals in response to abiotic stress. Given the current climate change and the urgency to balance our need for food and fuel, it is essential to understand how plants signal both, abiotic and biotic stresses not only locally but also to distal parts of the plant. In other words: to make plants that can survive adverse environments like drought, we need to know how plants move these lipophilic signals from root to shoot or vice versa.
Plant long-distance transport systems
Plants have evolved two long-distance transport systems: the xylem and the phloem:
The xylem moves water, minerals, and nutrients that were taken up from the soil throughout the plant. Transport occurs upwards through xylem vessels, which are dead cells with thickened peripheral cell walls, and is driven by the water potential gradient between the (wet) soil and the (dry) air. While some proteins have been found within this xylem stream [1, 2], no attempt has so far been made to identify lipophilic compounds.
The phloem is the main conduit for the transport of photoassimilates, the sugars produced through photosynthesis. It is composed of large tubular cells, known as sieve elements, which connect to form sieve tubes and can reach up to 100 m in tall trees. To allow for an unobstructed flow, the sieve elements, cells through which this transport occurs, have lost their nuclei, ribosomes, and most organelles during development. Cell walls at the interphase between two sieve elements contain large sieve pores [reviewed in 3]. Molecules found within the sieve element are thought to be synthesized in the neighboring companion cells and transported into the sieve element (SE) via plasmodesmata. Transport of photoassimilates as well as signaling molecules is thought to occur from source (photosynthetically active mature leaves) to sink (immature leaves, roots, fruits, flowers, etc.) in a mechanism driven by the osmotic gradient (“Münch’s Pressure flow hypothesis”; for a review see [3, 4]). Hence phloem transport can be bidirectional, moving up or down.
Messing with a dogma
Until less than 20 years ago, the phloem was thought of as nothing more than a transport system for photoassimilates. However, improvements in mass spectrometry techniques, the development of MALDI and electrospray ionization, and the increased sensitivity and resolution of mass spectrometers have allowed us to detect compounds present in only minute amounts. It is now accepted that the phloem is a complex trafficking system for stress signals and developmental regulators in the form of small molecules, peptides/proteins, and nucleic acids. A thorough summary of phloem contents, development, structure, and function can be found in [3]
More recently, complex lipids have also been detected in phloem exudates of several plant species [5-7]. Some were identified based on the activity of a fraction of phloem exudates, other identifications are based on a more comprehensive phloem lipid analysis. The questions are: What are these lipids? What is their function? And how are they solubilized and transported in the aqueous environment of the phloem?
Lipophilic molecules in the phloem
The lipophilic molecules found in the phloem can be clustered into the following groups (Table1):
Lipophilic hormones: Abscisic acid (ABA), Indole acetic acid (IAA), Gibberellins (GA), Cytokinins, Salicylic acid (SA), and oxylipins like Jasmonic acid (JA) and its precursor cis(+)-12-oxo phytodienoic acid (OPDA).
Small lipophilic molecules: Dehydroabietinal (DA), Azelaic acid (AzA), an unidentified Glycerol-3-phosphate derivative (G3P*), and fatty acids.
Steroid lipids: Free, acylated and glycosylated derivatives of cholesterol, sitosterol, camposterol and stigmasterol.
Glycerolipids: Diacylglyerol (DAG), Triacylglycerol (TAG), Phosphatidyl choline (PC), Phosphatidyl inositol (PI), Phosphatidic acid (PA), Monogalactosyl diacylglycerol (MGDG).

What is the lipid function in the phloem?
Lipids in the phloem could be found for a variety of reasons:
1) Phloem lipids could be products of membrane turnover or membrane damage [3]. When analyzing phloem lipids, the phospholipid profiles were found to be distinct from leaf lipids, with several lipids being unique to the phloem suggesting that membrane turnover in the phloem of healthy, intact leaves is unlikely [5]. However, membrane degradation as a result of injury or of a process similar to leaf senescence in the fall is a possibility. This process would begin with the lipolytic release of fatty acids from membrane lipids followed by β-oxidation in peroxisomes and conversion to sucrose [9]. As a result, it is not the lipid/fatty acid that is mobilized but the sucrose to which they had been converted. In addition, the caveat of this concept lies in the fact that neither peroxisomes nor enzymes for β-oxidation have so far been described in sieve elements, making this process within the sieve element rather unlikely.
2) Phloem lipids could be transported as building blocks or energy carriers. This is unlikely since most plant tissues are capable of lipid biosynthesis and thus, the need for lipids as precursors does not exist [10]. On the other hand, the phloem composition changes during leaf senescence from sucrose to amino acids as predominant molecules [11]. While this does not apply to the typical phloem sample obtained from young, healthy leaves, it is conceivable that fatty acids and triacylglycerols are moved as part of the seasonal senescence-related mobilization of cellular components. However, as mentioned above, this mobilization tends to be in the form of sucrose.
3) The third and most exciting option is that phloem lipids serve as long-distance signals. We already know that many lipids are important signaling compounds in plants! Yet despite finding and describing the function of some of the “lipid hormones” (ABA, IAA, GAs, JA, see [3, 12]) and small lipophilic molecules (DA, AzA; see [13]) in the phloem, their signaling function, particularly in case of phospholipids like PA, PI, and PC, and their transport mechanisms are only partially understood.
Movement of lipids in the aqueous environment of the phloem
Long-distance transport of hydrophobic compounds in aqueous systems like the phloem is not without precedence in biological systems: in animals lipids move cell-to-cell [14] or within the bloodstream [15, 16], often while bound to proteins. In addition, they also regulate gene expression [17, 18]. While these lipid-protein mechanisms are key to mammalian development and health, their possible importance in plants is virtually unexplored.
Small lipophilic molecules (lipophilic hormones, oxylipins, phytosterols, DA, and AzA) are already discussed as long-distance signals in the phloem. Phospholipids as long-distance signaling molecules provide a novel aspect in phloem-mediated signaling. Below are examples of the different mechanisms by which plants move lipids into and around the phloem, the lipid function, as well as protein candidates for specific functions in the movement of complex lipids:
Lipophilic hormones (for more details see reviews by [3, 12])
Auxins have been detected in the phloem of over 14 species. Auxin travels via polar auxin transport employing specific carriers for cell-to-cell movement across the membrane, as well as through bulk flow in the phloem. However, whether it is synthesized in the SEs or imported from other cells is as of yet unknown as is its movement mechanism in the phloem. It has been proposed that Auxin is moved into the phloem through carrier-mediated transport [19] employing the same transporters as during polar Auxin transport. Many Auxins in the phloem are esterified or, in at least one case, conjugated to the amino acid aspartic acid. This esterification could provide increased solubility and/or stability of the hormone.
A similar mechanism is employed in Jasmonic acid (JA) signaling. The oxylipin JA is probably the best-studied phloem lipid. It is synthesized in response to wounding or herbivory and moves throughout the plant to elicit a (systemic) defense response. This movement occurs in the form of JA-isoleucine [20]. Other researchers had shown movement of methyl-JA in the vasculature, a movement, which appeared to also extend to the xylem [21, 22]. Moreover, it has been suggested that the JA-precursor 12-Oxo-phytodienoic acid (OPDA), a second phloem-mobile oxylipin [23], may play a role in drought response and crosstalk with ABA. [24]. Salicylic acid like JA plays a pivotal role in systemic acquired resistance.
ABA as well as its metabolites phaseic acid and dihydrophaseic acid have been detected in the phloem of several plant species, yet no evidence could be found for glycosides or glucose esters. The presence of Gibberellin-like compounds in the phloem has been determined through bioassays of phloem sap as well as in the honeydew of phloem-sucking insects. GC-MS showed the presence of GA1 as well as several precursors but no metabolites. Its exact movement mechanism remains underdetermined. Cytokinins have been reported in the xylem and the phloem as early as the 1960s. The major form of Cytokinin in the xylem is trans-zeatin riboside, while N6-(2-isopentenyl) adenine-type Cytokinin-ribosides are the predominant forms in the phloem (for a review see [25]). Translocation in the xylem is controlled by environmental factors with trans-zeatin riboside as a signal for the nitrate status. Thus, it is conceivable that the chemical form of the Cytokinin present determines not only the mode and direction of its transport but also its signaling function. Movement of Cytokinins (as well as Auxin) through the phloem has been shown by using grafting as well as feeding with radiolabeled compounds.
While the lipophilic hormones are very diverse in structure, they all appear to move through the phloem either in their free form or as conjugates with amino acids, methyl groups or sugar (glycosyl-/ ribosyl-) groups. At this point, (carrier-) proteins do not appear to be necessary for their long-distance movement.
Small lipophilic molecules
Small lipophilic molecules found in the phloem are Dehydroabietinal (DA), Azelaic acid (AzA), an unidentified Glycerol-3-phosphate derivative (G3P*), and fatty acids. They play a crucial role in systemic acquired resistance. Yet, their ability to move and the mechanisms, by which this occurs, remains to be elucidated [see 13].
Steroid lipids
Behmer et al. [7] detected free, acylated and glycosylated derivatives of cholesterol, sitosterol, camposterol, and stigmasterol in the phloem. They found that about half of the phloem sterol pool was glycosylated while the remainder consisted of free- and acylated forms. While these findings suggest that their transport in the phloem may be dependent on pressure-flow based movement of free, acylated or glycosylated sterols, the exact mechanism remains to be determined but might occur via as of yet unidentified lipid-transport proteins [7].
Phospholipids
Lipids, specifically phospholipids, are mainly found within cell membranes for which they provide structure but also act as mediators regulating various aspects of plant development and environmental interactions [26]. Several of these lipids including PA, LPA (lysophosphatidic acid), DAG, PI and its phosphates (PIPs) can function as second messengers within plant cells. However, virtually nothing is known about their possible function in long-distance signaling. Unlike structural lipids, signaling lipids are present only in minute amounts. They accumulate rapidly and transiently in response to external stimuli. We and others have identified several polar lipids in phloem exudates of Arabidopsis [5] and canola [6], among them the known intracellular signaling molecules/precursors PA, PI, and PC. In the leaf, PA is produced in response to several abiotic stresses like drought, salinity, wounding, and cold, but also as a consequence of biotic stress like pathogen infection and its signals like oxylipins [26]. It is generated by either the PLC-DGK pathway or directly by PLD, depending on the stress [26, 27]. The presence of phospholipids and in particular, PA, prompted us to suggest that phloem phospholipids could act as long-distance signals as well. Due to their hydrophobic nature, we proposed that phloem lipid-binding proteins participate in different aspects of this signaling cascade ([8, 28]; see Figure 1; from [29]:
These proteins: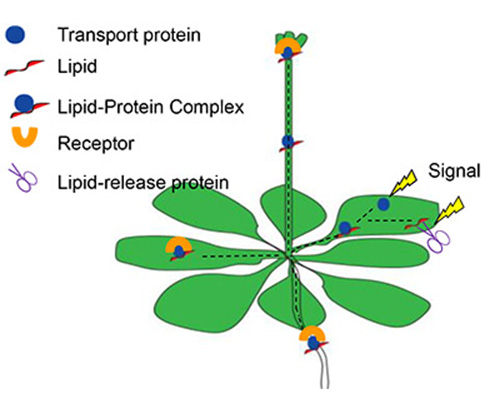
(i) could move or release the lipid into the phloem,
(ii) solubilize the lipid and transport it to its target organ,
(iii) function as part of a receptor that binds the signaling lipid and affects development.
Several currently studied phloem-localized lipid-binding proteins could function in translocation aspects of the long-distance lipid-signaling mechanism proposed in figure 1 (see also 8):
Flowering-locus T (FT; rice homologue: Hd3a)is a phloem-mobile lipid-binding protein shown to be involved in long-distance signaling of flowering and, at least in the case of the poplar FT, in signaling seasonal leaf abscission as well. Recent evidence suggests that AtFT binds PC [30]. While PC has been found in phloem exudates, it remains to be investigated whether the PC-Ft interaction occurs in mesophyll, SE or apical meristem cells and whether PC is part of the mobile signal.
A second lipid-binding protein with signaling function is Dir1, a lipid-transfer protein that is associated with SAR. Locally-induced Dir1 can travel to distal leaves and binds lysophosphatidyl choline (LPC; [31, 32]). However, it has also been detected in a high-molecular weight complex, which would not allow for long-distance movement but may have an alternative signaling function [13]. Hence, whether the LPC-Dir1 complex functions as signal or as a receptor complex continues to be unclear.
ACBP6 is a small acyl-CoA binding protein that is expressed in the vasculature and binds 18:3 fatty acids (α-linolenic acid/ALA). ALA is the precursor fatty acid for JA biosynthesis. In this case it could be transported as an important nutritional component or as a signaling molecule/precursor for JA biosynthesis. It was suggested that ACBP6 could facilitate movement of ALA into the phloem [27].
PLAFP is a 20kDa PA-binding protein. Both, protein and lipid were found in the phloem and are induced in response to several abiotic stresses. Yet their mobility remains to be shown. Hence, as with the previous lipid-protein complexes, the mechanism of the protein-lipid interaction in long-distance signaling remains unclear. Modeling of the lipid-protein complex, suggests however the possibility of PA binding into a hydrophobic groove in PLAFP, thus sequestering it away from the aqueous environment and allowing for the possibility of movement. If in any of these cases, the protein modulates the phloem lipid content and a joint movement can be shown, this would open a completely new era of phospholipid signaling.
Acknowledgements
This work was supported by the National Science Foundation (NSF-IOS #1144391) and the United States Department of Agriculture (USDA-NIFA Hatch project #MICL02233)
References
- Dafoe, N.J. and Constabel, C.P. Proteomic analysis of hybrid poplar xylem sap. Phytochemistry. 70, 856-63 (2009) (doi: 10.1016/j.phytochem.2009.04.016).
- de Bernonville, T.D., Albenne, C., Arlat, M., Hoffmann, L., Lauber, E. and Jamet, E. Xylem sap proteomics. Methods Mol. Biol. 1072, 391-405 (2014) (doi: 10.1007/978-1-62703-631-3_28).
- Lucas, W.J., Groover, A., Lichtenberger, R., Furuta, K., Yadav, S.R., Helariutta, Y., He, X.Q., Fukuda, H., Kang, J., Brady, S.M., Patrick, J.W., Sperry, J., Yoshida, A., López-Millán, A.F., Grusak, M.A. and Kachroo, P. The plant vascular system: evolution, development and functions. J. Integr. Plant. Biol. 55, 294-388 (2013).
- Froehlich, D.R., Mullendore, D.L., Jensen, K.H., Ross-Elliott, T.J., Anstead, J.A., Thompson, G.A., Pélissier, H.C. and Knoblauch, M. Phloem ultrastructure and pressure flow: Sieve-element-occlusion-related agglomerations do not affect translocation. Plant Cell 23, 4428–4445 (2011).
- Guelette, B.S., Benning, U.F. and Hoffmann-Benning, S. Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 63, 3603 (2012).
- Madey, E., Nowack, L.M. and Thompson, J.E. Isolation and characterization of lipid in phloem sap of canola. Planta 214, 625-634 (2002).
- Behmer, S.T., Olszewski, N., Sebastiani, J., Palka, S., Sparacino, G., Sciarrno, E., and Grebenok, R.J. Plant phloem sterol content: forms, putative functions, and implications for phloem-feeding insects Front. Plant Sci. 4, 370 (2013).
- Benning, U.F., Tamot, B., Guelette, B.S., Hoffmann-Benning, S. New aspects of phloem mediated long-distance lipid signaling in plants. Front. Plant Sci. 3: 53 (2012).
- Troncoso-Ponce, M.A., Cao, X., Yang, Z., Ohlrogge, J.B. Lipid turnover during senescence. Plant Science Volumes 205–206, 13–19 (2013).
- Ohlrogge, J. and Browse, J. Lipid Biosynthesis. The Plant Cell 7, 957-970 (1995).
- Thomas, H. and Stoddard, J.L. Leaf Senescence. Annu. Rev.Plant Physiol. 31, 83-111 (1980) (DOI: 10.1146/annurev.pp.31.060180.000503).
- Vian , A., Stankovic, B. and Davies, E. Signalomics: Diversity and Methods of Analysis of Systemic Signals in Plants. In: PlantOmics: The Omics of Plant Science pp. 459-489. (D. Barh, M.S. Khanand E. Davies (eds.), Springer Books) (2015).
- Shah, J., Chaturvedi, R., Chowdhury, Z., Venables, B. and Petros, R.A. Signaling by small metabolites in systemic acquired Resistance. The Plant Journal 79, 645–658 (2014).
- Christie, W.W. http://lipidlibrary.aocs.org/content.cfm?ItemNumber=39350 (2014).
- Charbonneau, D., Beauregard, M. and Tajmir-Riahi, H.-A. Structural Analysis of Human Serum Albumin Complexes with Cationic Lipids. The Journal of physical chemistry B 113, 1777-1781 (2009).
- Blaner, W.S. Retinol-Binding Protein: The Serum Transport Protein for Vitamin A. Endocrine Reviews 10, 308-316 (1989).
- Musille, P.M., Kohn, J.A. and Ortlund, E.A. Phospholipid-driven gene regulation. FEBS Lett. 587, 1238-46 (2013).
- Wahli, W. and Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends in Endocrinology and Metabolism 23, 351-363 (2012)
- Petrášek, J. and Friml, J. Auxin transport routes in plant development. Development 136, 2675-2688 (2009) (Doi:10.1242/dev.030353).
- Matsuura, H., Takeishi, S., Kiatoka, N., Sato, C., Sueda, K., Masuta, C. and Nabeta, K. Transportation of de novo synthesized jasmonoyl isoleucine in tomato. Phytochemistry 83, 25–33 (2012).
- Tamogami, S., Noge, K., Abe, M., Agrawal, G.K. and Rakwal, R. Methyl jasmonate is transported to distal leaves via vascular process metabolizing itself into JA-Ile and triggering VOCs emission as defensive metabolites. Plant Signal. Behav. 7, 1378-81 (2012) (Doi: 10.4161/psb.21762).
- Thorpe, M.R., Ferrieri, A.P., Herth, M.M., and Ferrieri, R.A. (11)C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226, 541-551 (2007).
- Landgraf P, Feussner I, Hunger A, Scheel D, Rosahl S Systemic accumulation of 12-oxo-phytodienoic acid in SAR-induced potato plants. Eur. J. Plant Pathol. 108, 279-283 (2001).
- Savchenko, T., Kolla, V.A., Wang, C.Q., Nasafi, Z., Hicks, D.R., Phadungchob, B., Chehab, W.E., Brandizzi, F., Froehlich, J., Dehesh, K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 164, 1151-1160 (2014).
- Kudo, T., Kiba, T. and Sakakibara, H. Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52, 53–60 (2010).
- Munnik, T. and Testerink, C. Plant phospholipid signaling: “in a nutshell”. J. Lipid Res. S260-S265 (2009).
- Wang, X., Guo, L., Wang, G., and Li, M. PLD: phospholipase Ds in plant signaling. In: Phospholipases in Plant Signaling, pp. 3-26 (Wang, X., ed., Springer books., 2014)
- Zheng, S.X., Xiao, S., and Chye, M.L. The gene encoding Arabidopsis Acyl-CoA-Binding Protein 3 is pathogen-inducible and subject to circadian regulation. J. Exp. Bot. 63, 2985-3000 (2012).
- bmb.natsci.msu.edu/faculty/susanne-hoffmann-benning-assistant-professor/current-research/
- Nakamura, Y., Andrés, F., Kanehara, K,, Liu, Y.C., Dörmann, P., and Coupland, G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nature Communications 5, 3553 (2014).
- Champigny, M.J., Isaacs, M., Carella, P., Faubert, J., Fobert, P.R. and Cameron, R.K. Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 4, 230 (2013).
- Lascombe, M.B., Bakan, B., Buhot, N., Marion, D., Blein, J.P., Larue, V., Lamb, C. and Prangé, T. The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci. 17, 1522-30 (2008).
- Figures were modified in part from Wikimedia commons: https://commons.wikimedia.org/
In This Section
- Plant Fatty Acid Synthesis
- Production of Unusual Fatty Acids in Plants
- Arabidopsis Acyl-Coenzyme A-Binding Proteins
- Long Chain acyl-coA Synthetases and Other Acyl Activating Enzymes
- Plant Triacylglycerol Synthesis
- Triacylglycerol Biosynthesis in Eukaryotic Microalgae
- Subcellular Oil Droplets and Oleosins in Plants
- Triacylglycerol Mobilisation in Plants
- Role of Transcription Factors in Storage Lipid Accumulation in Plants
- Biosynthesis of Plant Lipid Polyesters
- Rubber Biosynthesis in Plants
- Carotenoid Biosynthesis and Regulation in Plants
- The Oxylipin Biosynthetic Pathways in Plants
- N-Acylphosphatidylethanolamines (NAPEs), N-acylethanolamines (NAEs) and Other Acylamides: Metabolism, Occurrence and Functions in Plants
- Phosphoinositide Signaling in Plants
- Plant Lipidomics
- 50 years of Galactolipid Research: The Beginnings
- Transport and function of lipids in the plant phloem
